Project
DANIO-CODE
Navigation
Downloads
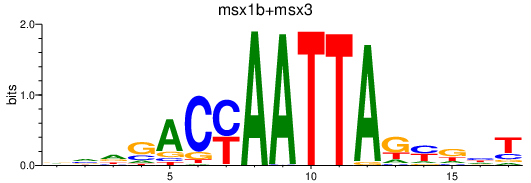
Results for msx1b+msx3
Z-value: 0.36
Transcription factors associated with msx1b+msx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx1b
|
ENSDARG00000008886 | muscle segment homeobox 1b |
|
msx3
|
ENSDARG00000015674 | muscle segment homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx1b | dr10_dc_chr1_+_48708504_48708540 | -0.26 | 3.3e-01 | Click! |
| msx3 | dr10_dc_chr13_+_24531753_24531851 | -0.11 | 6.8e-01 | Click! |
Activity profile of msx1b+msx3 motif
Sorted Z-values of msx1b+msx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of msx1b+msx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_4825461 | 0.35 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr14_+_34150130 | 0.32 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr12_-_33256671 | 0.30 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr17_-_45021393 | 0.27 |
|
|
|
| chr10_+_17277353 | 0.26 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr5_+_19429620 | 0.25 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr5_+_19429500 | 0.23 |
ENSDART00000168868
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr13_-_18564182 | 0.22 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr12_-_33256754 | 0.22 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_-_32532691 | 0.21 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr12_-_33256599 | 0.20 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr24_-_7928026 | 0.18 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr19_-_18664720 | 0.18 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr4_-_75683584 | 0.17 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr20_-_51454257 | 0.17 |
ENSDART00000023064
|
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr12_-_33257026 | 0.17 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr19_+_9066225 | 0.16 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr4_-_62022037 | 0.16 |
|
|
|
| chr25_-_25455861 | 0.16 |
ENSDART00000150412
|
irf7
|
interferon regulatory factor 7 |
| chr9_-_32532843 | 0.15 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr6_-_9459739 | 0.14 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr2_+_6341404 | 0.14 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr19_-_18664670 | 0.14 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr2_+_6341345 | 0.14 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr18_-_43890836 | 0.14 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr22_-_26254136 | 0.13 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr5_+_29193876 | 0.13 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr24_-_24999240 | 0.13 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr7_+_23799153 | 0.13 |
|
|
|
| chr17_-_35331055 | 0.12 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr20_-_51454057 | 0.12 |
ENSDART00000023064
|
slc35b2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr21_-_20291707 | 0.11 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr2_+_31974269 | 0.11 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| KN150663v1_-_3381 | 0.11 |
|
|
|
| chr12_-_33256934 | 0.11 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr16_+_41222034 | 0.11 |
ENSDART00000135294
|
nek11
|
NIMA-related kinase 11 |
| chr24_-_24859334 | 0.10 |
ENSDART00000080997
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr10_-_34927807 | 0.10 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr3_-_26060787 | 0.10 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr4_+_4825409 | 0.10 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr21_-_20291586 | 0.10 |
ENSDART00000079593
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr15_+_30580253 | 0.10 |
|
|
|
| chr2_-_20490037 | 0.09 |
ENSDART00000160388
|
FQ377605.1
|
ENSDARG00000101927 |
| chr8_-_45830321 | 0.09 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr10_+_39291358 | 0.09 |
ENSDART00000159501
|
stt3a
|
STT3A, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr4_+_4825628 | 0.09 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr5_-_19429016 | 0.09 |
ENSDART00000170344
|
git2a
|
G protein-coupled receptor kinase interacting ArfGAP 2a |
| chr16_+_9822930 | 0.09 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr2_+_1260407 | 0.09 |
|
|
|
| chr16_-_25653129 | 0.09 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr5_+_58327436 | 0.08 |
ENSDART00000062175
|
derl2
|
derlin 2 |
| chr6_+_50452131 | 0.08 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr17_-_35331124 | 0.08 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr12_+_33257120 | 0.08 |
|
|
|
| chr16_+_41222408 | 0.07 |
ENSDART00000128963
|
nek11
|
NIMA-related kinase 11 |
| chr14_-_46649324 | 0.07 |
|
|
|
| chr24_-_7928168 | 0.06 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr14_-_46649231 | 0.06 |
|
|
|
| chr23_-_35384196 | 0.06 |
ENSDART00000138660
|
fbxo25
|
F-box protein 25 |
| chr12_-_28145021 | 0.06 |
|
|
|
| chr5_+_62987426 | 0.06 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr20_+_28901045 | 0.06 |
ENSDART00000153046
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr7_-_8128948 | 0.05 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr13_-_36672606 | 0.05 |
ENSDART00000179242
|
sav1
|
salvador family WW domain containing protein 1 |
| chr17_-_35330764 | 0.05 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr15_-_5827067 | 0.05 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr20_-_19611724 | 0.05 |
ENSDART00000168521
|
snx17
|
sorting nexin 17 |
| chr20_+_1724609 | 0.05 |
|
|
|
| chr25_-_12236333 | 0.05 |
ENSDART00000174863
ENSDART00000179042 |
BX323452.1
|
ENSDARG00000107774 |
| chr11_-_1939192 | 0.04 |
ENSDART00000172885
|
faim2b
|
Fas apoptotic inhibitory molecule 2b |
| chr14_-_46649718 | 0.04 |
|
|
|
| chr22_+_12746080 | 0.04 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr7_+_32451041 | 0.04 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr11_-_11925832 | 0.04 |
|
|
|
| chr18_-_17527155 | 0.04 |
|
|
|
| chr8_+_20108592 | 0.03 |
|
|
|
| chr24_-_24999348 | 0.03 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr20_+_1724656 | 0.03 |
|
|
|
| chr11_+_24479085 | 0.03 |
ENSDART00000145217
|
ENSDARG00000070571
|
ENSDARG00000070571 |
| chr14_+_34150232 | 0.03 |
ENSDART00000148044
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr19_-_8849482 | 0.03 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr1_+_11541423 | 0.02 |
ENSDART00000170760
|
tmod1
|
tropomodulin 1 |
| chr7_+_59371957 | 0.02 |
ENSDART00000125570
|
trmt44
|
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr1_-_45941709 | 0.02 |
ENSDART00000053232
|
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr8_-_17786546 | 0.02 |
ENSDART00000063587
|
st6galnac5b
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5b |
| chr2_+_45447068 | 0.02 |
ENSDART00000083957
ENSDART00000160867 |
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr17_+_41126669 | 0.02 |
ENSDART00000137091
|
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr12_+_3043444 | 0.02 |
ENSDART00000149427
|
sgca
|
sarcoglycan, alpha |
| chr5_-_47106694 | 0.02 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr14_+_20621407 | 0.02 |
ENSDART00000144736
|
zgc:66433
|
zgc:66433 |
| chr1_-_18809429 | 0.02 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr15_-_42081757 | 0.02 |
ENSDART00000004338
|
epha4l
|
eph receptor A4, like |
| chr24_-_24859135 | 0.02 |
ENSDART00000136860
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr21_+_6829161 | 0.01 |
ENSDART00000037265
|
olfm1b
|
olfactomedin 1b |
| chr11_-_40192951 | 0.01 |
ENSDART00000102750
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr8_-_53850837 | 0.01 |
ENSDART00000172249
|
wnt5a
|
wingless-type MMTV integration site family, member 5a |
| chr14_-_46648801 | 0.01 |
|
|
|
| chr2_-_3846844 | 0.01 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr14_+_31596468 | 0.01 |
ENSDART00000173259
|
BX005454.1
|
ENSDARG00000100646 |
| chr17_-_41126450 | 0.01 |
ENSDART00000030622
|
rbks
|
ribokinase |
| chr1_-_10373506 | 0.01 |
|
|
|
| chr21_+_6829352 | 0.01 |
ENSDART00000146371
|
olfm1b
|
olfactomedin 1b |
| chr25_-_27120954 | 0.00 |
ENSDART00000157319
|
hyal4
|
hyaluronoglucosaminidase 4 |
| chr20_+_25669615 | 0.00 |
ENSDART00000063107
|
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr17_+_32547862 | 0.00 |
ENSDART00000018423
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr23_-_35384257 | 0.00 |
ENSDART00000113643
|
fbxo25
|
F-box protein 25 |
| chr5_+_31590746 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.0 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.3 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |