Project
DANIO-CODE
Navigation
Downloads
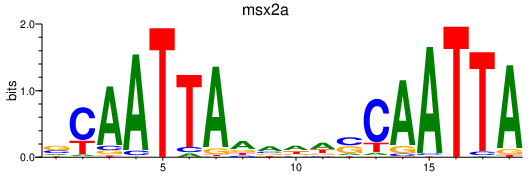
Results for msx2a
Z-value: 0.33
Transcription factors associated with msx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2a
|
ENSDARG00000104651 | muscle segment homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2a | dr10_dc_chr14_-_23784679_23784711 | -0.75 | 8.1e-04 | Click! |
Activity profile of msx2a motif
Sorted Z-values of msx2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_868378 | 1.14 |
|
|
|
| chr13_-_18564182 | 0.71 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr19_+_31997954 | 0.59 |
ENSDART00000164108
|
gmnn
|
geminin, DNA replication inhibitor |
| chr13_+_38100228 | 0.56 |
ENSDART00000144093
|
si:dkeyp-4c7.3
|
si:dkeyp-4c7.3 |
| chr3_-_15346590 | 0.47 |
ENSDART00000124063
|
sgf29
|
SAGA complex associated factor 29 |
| chr17_+_23710004 | 0.46 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr4_-_6407602 | 0.45 |
ENSDART00000134376
|
CR356233.2
|
ENSDARG00000093020 |
| chr11_+_17849608 | 0.44 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr7_-_51497945 | 0.43 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr17_+_23709781 | 0.41 |
ENSDART00000034913
|
zgc:91976
|
zgc:91976 |
| chr5_-_18458903 | 0.40 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr21_+_5027444 | 0.40 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr21_-_2292663 | 0.40 |
ENSDART00000164015
|
zgc:66483
|
zgc:66483 |
| chr17_+_23709676 | 0.39 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr6_+_21887963 | 0.39 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr25_+_32716795 | 0.39 |
ENSDART00000131098
|
ENSDARG00000087402
|
ENSDARG00000087402 |
| chr3_-_15346427 | 0.37 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr14_+_34673281 | 0.37 |
ENSDART00000164974
|
ebf1a
|
early B-cell factor 1a |
| chr6_+_21887935 | 0.35 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr20_+_36957917 | 0.33 |
ENSDART00000153091
|
BX248390.1
|
ENSDARG00000096735 |
| chr12_+_32316774 | 0.33 |
ENSDART00000153144
|
CT573006.1
|
ENSDARG00000096761 |
| chr3_-_33811737 | 0.32 |
ENSDART00000026090
ENSDART00000047660 ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr7_-_20322448 | 0.32 |
ENSDART00000170850
|
CR318653.1
|
ENSDARG00000104557 |
| chr16_-_13723352 | 0.31 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr9_-_40129231 | 0.31 |
ENSDART00000177531
|
CABZ01092969.1
|
ENSDARG00000108990 |
| chr15_-_23786063 | 0.30 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr21_+_5027623 | 0.30 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr3_-_28371648 | 0.30 |
ENSDART00000150912
|
CR792426.1
|
ENSDARG00000096347 |
| chr22_+_2735606 | 0.30 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr25_+_18997440 | 0.30 |
ENSDART00000155569
|
si:dkeyp-19h3.8
|
si:dkeyp-19h3.8 |
| chr12_-_25126122 | 0.29 |
ENSDART00000152931
|
kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr21_+_5027668 | 0.29 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr7_+_24257251 | 0.29 |
ENSDART00000136473
|
ENSDARG00000079281
|
ENSDARG00000079281 |
| chr5_-_19619115 | 0.29 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr3_-_26675055 | 0.28 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr3_+_27582277 | 0.28 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr19_-_5186692 | 0.27 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr5_-_19619201 | 0.26 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr2_+_1260407 | 0.26 |
|
|
|
| chr23_+_16848278 | 0.26 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr4_-_14193396 | 0.26 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr4_-_75683584 | 0.26 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr14_+_29601073 | 0.25 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr7_+_39392939 | 0.25 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr19_-_27093002 | 0.25 |
|
|
|
| chr5_-_67839187 | 0.24 |
|
|
|
| chr5_+_27793171 | 0.24 |
|
|
|
| chr21_-_13654658 | 0.23 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr5_+_71100066 | 0.23 |
ENSDART00000115182
|
nup214
|
nucleoporin 214 |
| chr16_-_13723295 | 0.22 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr15_-_6969345 | 0.21 |
ENSDART00000169529
|
mrps22
|
mitochondrial ribosomal protein S22 |
| chr5_+_21673232 | 0.21 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr3_-_15346710 | 0.20 |
ENSDART00000007726
|
sgf29
|
SAGA complex associated factor 29 |
| chr3_+_32278978 | 0.19 |
ENSDART00000061426
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr7_-_20322390 | 0.19 |
ENSDART00000170850
|
CR318653.1
|
ENSDARG00000104557 |
| chr22_+_37696341 | 0.19 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr21_-_39616469 | 0.19 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr13_-_40592360 | 0.19 |
|
|
|
| chr24_-_32668102 | 0.19 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr5_-_39077051 | 0.19 |
|
|
|
| chr14_+_17092009 | 0.19 |
ENSDART00000129838
|
rnf212
|
ring finger protein 212 |
| chr3_-_58094618 | 0.19 |
ENSDART00000093031
|
snu13a
|
SNU13 homolog, small nuclear ribonucleoprotein a (U4/U6.U5) |
| chr16_+_1843899 | 0.19 |
|
|
|
| chr10_+_35209240 | 0.19 |
ENSDART00000126105
|
nsun5
|
NOP2/Sun domain family, member 5 |
| chr25_-_36678422 | 0.18 |
ENSDART00000087247
|
glg1a
|
golgi glycoprotein 1a |
| chr3_-_25361866 | 0.17 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr15_+_2556689 | 0.17 |
ENSDART00000063329
|
cux1b
|
cut-like homeobox 1b |
| chr10_-_42279805 | 0.17 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr23_+_44937093 | 0.17 |
|
|
|
| chr11_-_25495239 | 0.17 |
|
|
|
| chr23_+_16848213 | 0.16 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr5_-_18458827 | 0.16 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr23_+_2847589 | 0.16 |
ENSDART00000156954
|
plcg1
|
phospholipase C, gamma 1 |
| chr21_+_5027576 | 0.16 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr9_-_32240567 | 0.16 |
|
|
|
| chr11_-_7786415 | 0.16 |
ENSDART00000154569
|
CR450813.1
|
ENSDARG00000097035 |
| chr1_+_29027527 | 0.16 |
ENSDART00000142184
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr3_-_33812379 | 0.15 |
|
|
|
| chr19_-_7522951 | 0.15 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr14_+_29601046 | 0.14 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr9_+_55963230 | 0.14 |
|
|
|
| chr19_-_11397220 | 0.14 |
ENSDART00000104933
|
eepd1
|
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr6_-_1638631 | 0.13 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr25_-_34596585 | 0.13 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr3_-_35411760 | 0.13 |
ENSDART00000022147
|
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr21_+_32046764 | 0.13 |
|
|
|
| chr2_-_41713521 | 0.13 |
ENSDART00000084597
|
d2hgdh
|
D-2-hydroxyglutarate dehydrogenase |
| chr10_-_42691422 | 0.12 |
ENSDART00000039187
|
march5l
|
membrane-associated ring finger (C3HC4) 5, like |
| chr1_-_19072200 | 0.12 |
|
|
|
| chr1_+_34741292 | 0.12 |
ENSDART00000109678
|
usp38
|
ubiquitin specific peptidase 38 |
| chr1_+_19071918 | 0.12 |
ENSDART00000138276
|
ENSDARG00000077648
|
ENSDARG00000077648 |
| chr25_+_20021806 | 0.11 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr14_+_29601252 | 0.11 |
ENSDART00000143763
|
fam149a
|
family with sequence similarity 149 member A |
| chr6_+_54798741 | 0.11 |
|
|
|
| chr18_-_199009 | 0.11 |
|
|
|
| chr1_+_9633924 | 0.11 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr25_-_34596670 | 0.11 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr21_+_812425 | 0.11 |
ENSDART00000006419
ENSDART00000133976 |
txnl1
|
thioredoxin-like 1 |
| chr16_-_26798351 | 0.11 |
|
|
|
| chr16_+_34157948 | 0.11 |
ENSDART00000140552
|
tcea3
|
transcription elongation factor A (SII), 3 |
| chr22_-_29938022 | 0.10 |
|
|
|
| chr22_-_6772249 | 0.10 |
ENSDART00000108883
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr19_-_27092958 | 0.10 |
|
|
|
| chr11_+_17849528 | 0.10 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_-_19666829 | 0.10 |
ENSDART00000083085
|
mtmr14
|
myotubularin related protein 14 |
| chr10_-_22181059 | 0.10 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr17_-_43896998 | 0.10 |
ENSDART00000156513
|
BX465227.1
|
ENSDARG00000097825 |
| chr3_-_25361708 | 0.10 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr16_+_28643677 | 0.10 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr10_-_5846612 | 0.10 |
ENSDART00000161096
|
ankrd55
|
ankyrin repeat domain 55 |
| chr23_-_29579049 | 0.10 |
ENSDART00000165744
|
kif1b
|
kinesin family member 1B |
| chr10_-_42279667 | 0.09 |
ENSDART00000132976
|
ess2
|
ess-2 splicing factor homolog |
| chr4_-_18976343 | 0.09 |
|
|
|
| chr22_-_27252706 | 0.09 |
ENSDART00000019442
|
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr16_+_11027467 | 0.09 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr7_+_22540012 | 0.09 |
|
|
|
| chr21_+_27152225 | 0.08 |
ENSDART00000125349
|
bada
|
BCL2-associated agonist of cell death a |
| chr19_+_40792676 | 0.08 |
ENSDART00000110699
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr5_+_53229583 | 0.08 |
ENSDART00000171107
|
naa38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit |
| chr2_-_58230370 | 0.08 |
|
|
|
| chr5_+_58883486 | 0.08 |
ENSDART00000055350
|
dtx2
|
deltex 2, E3 ubiquitin ligase |
| chr11_+_42184011 | 0.08 |
ENSDART00000161332
|
zgc:110286
|
zgc:110286 |
| chr12_+_33793468 | 0.08 |
ENSDART00000130853
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr8_-_53553764 | 0.08 |
ENSDART00000157521
|
actr8
|
ARP8 actin related protein 8 homolog |
| chr6_-_32059004 | 0.08 |
ENSDART00000139055
|
efcab7
|
EF-hand calcium binding domain 7 |
| chr10_+_3053045 | 0.08 |
ENSDART00000028870
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr1_+_9634016 | 0.07 |
ENSDART00000029774
|
tmem55bb
|
transmembrane protein 55Bb |
| chr7_+_39393052 | 0.07 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr2_+_1260457 | 0.07 |
|
|
|
| chr3_-_25361754 | 0.07 |
ENSDART00000147322
|
grb2b
|
growth factor receptor-bound protein 2b |
| chr25_+_18997277 | 0.06 |
ENSDART00000154066
|
isg20
|
interferon stimulated exonuclease gene |
| chr11_-_16017396 | 0.06 |
ENSDART00000081062
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr22_-_6772204 | 0.06 |
ENSDART00000108883
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr24_+_20782714 | 0.06 |
ENSDART00000144883
|
fam162a
|
family with sequence similarity 162, member A |
| chr16_+_12726556 | 0.06 |
|
|
|
| chr19_+_40792612 | 0.05 |
ENSDART00000017917
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr7_+_39393106 | 0.05 |
ENSDART00000047271
ENSDART00000113458 |
zgc:158564
|
zgc:158564 |
| chr13_-_5952897 | 0.04 |
ENSDART00000099224
|
dld
|
deltaD |
| chr14_-_43204831 | 0.04 |
ENSDART00000122339
|
gar1
|
GAR1 homolog, ribonucleoprotein |
| chr11_+_42184049 | 0.04 |
ENSDART00000056048
|
si:ch1073-165f9.2
|
si:ch1073-165f9.2 |
| chr1_-_44513922 | 0.03 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr16_-_21891328 | 0.03 |
ENSDART00000123597
|
ENSDARG00000089032
|
ENSDARG00000089032 |
| chr6_-_55332828 | 0.03 |
|
|
|
| chr13_+_40560230 | 0.03 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr11_+_3939876 | 0.03 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr17_+_30431395 | 0.03 |
ENSDART00000153939
|
lpin1
|
lipin 1 |
| chr2_+_48220021 | 0.03 |
ENSDART00000056291
|
klf6b
|
Kruppel-like factor 6b |
| chr21_+_16969092 | 0.03 |
ENSDART00000101246
|
vps29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr4_-_9195261 | 0.02 |
ENSDART00000080943
|
hcfc2
|
host cell factor C2 |
| chr16_-_36794503 | 0.02 |
ENSDART00000133310
|
pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr10_-_42691305 | 0.02 |
ENSDART00000171404
|
march5l
|
membrane-associated ring finger (C3HC4) 5, like |
| chr22_+_4408184 | 0.02 |
ENSDART00000113811
|
CU570881.1
|
ENSDARG00000078024 |
| chr14_-_33604914 | 0.01 |
ENSDART00000168546
|
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr7_-_26243985 | 0.01 |
ENSDART00000173617
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr6_-_1638559 | 0.01 |
ENSDART00000087039
|
ENSDARG00000015563
|
ENSDARG00000015563 |
| chr12_-_6029653 | 0.01 |
ENSDART00000002583
|
aarsd1
|
alanyl-tRNA synthetase domain containing 1 |
| chr2_+_2583038 | 0.01 |
|
|
|
| chr15_+_2556736 | 0.01 |
ENSDART00000063329
|
cux1b
|
cut-like homeobox 1b |
| chr12_-_29190576 | 0.01 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr19_-_30767032 | 0.01 |
ENSDART00000146847
|
khdrbs1b
|
KH domain containing, RNA binding, signal transduction associated 1b |
| chr1_+_264808 | 0.01 |
ENSDART00000139438
|
cenpe
|
centromere protein E |
| chr2_-_57723042 | 0.00 |
ENSDART00000143973
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr3_+_59589031 | 0.00 |
ENSDART00000108647
|
alyref
|
Aly/REF export factor |
| chr13_-_743812 | 0.00 |
ENSDART00000167256
|
ENSDARG00000102651
|
ENSDARG00000102651 |
| chr6_-_6965908 | 0.00 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr14_-_46646688 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0022602 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 0.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.2 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.1 | 0.2 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.1 | GO:0090344 | negative regulation of cell aging(GO:0090344) |
| 0.0 | 1.0 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0045199 | maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.7 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.0 | GO:1903232 | melanosome assembly(GO:1903232) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0031085 | BLOC-3 complex(GO:0031085) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.7 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |