Project
DANIO-CODE
Navigation
Downloads
Results for msx2b
Z-value: 0.83
Transcription factors associated with msx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2b
|
ENSDARG00000101023 | muscle segment homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2b | dr10_dc_chr21_-_41286846_41286877 | -0.85 | 3.0e-05 | Click! |
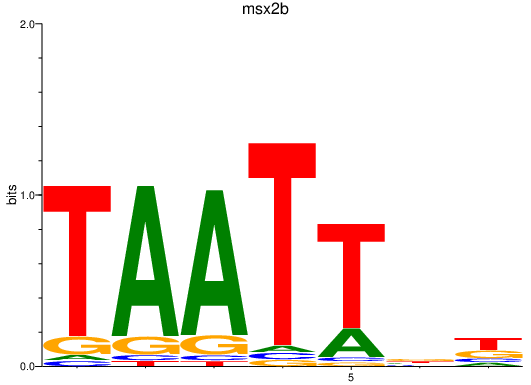
Activity profile of msx2b motif
Sorted Z-values of msx2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_49934440 | 3.51 |
ENSDART00000034541
|
gpatch11
|
G patch domain containing 11 |
| chr10_-_25246786 | 2.84 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr14_-_880799 | 2.59 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr5_-_8712068 | 2.19 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr16_-_24727689 | 1.95 |
ENSDART00000167121
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr5_-_8712114 | 1.73 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr19_-_48327666 | 1.71 |
|
|
|
| chr13_+_33331767 | 1.59 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr20_-_3220106 | 1.45 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr7_+_16771283 | 1.44 |
ENSDART00000171275
|
CU929054.1
|
ENSDARG00000102605 |
| chr2_-_37418967 | 1.41 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr10_+_11014479 | 1.36 |
ENSDART00000064992
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr19_+_14132374 | 1.31 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr17_+_50622734 | 1.28 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr20_-_23527234 | 1.28 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr11_+_25302044 | 1.23 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr18_+_6417959 | 1.21 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr14_+_33167424 | 1.20 |
ENSDART00000038128
|
mcts1
|
malignant T cell amplified sequence 1 |
| chr2_+_46210971 | 1.19 |
|
|
|
| chr2_+_46210909 | 1.13 |
|
|
|
| chr22_+_4734033 | 1.08 |
ENSDART00000158846
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr15_-_23786063 | 1.08 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr21_-_37750338 | 1.07 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr12_-_17741310 | 1.05 |
ENSDART00000166604
|
baiap2l1a
|
BAI1-associated protein 2-like 1a |
| chr17_+_32670368 | 1.05 |
ENSDART00000142449
|
ctsba
|
cathepsin Ba |
| KN149710v1_+_38638 | 1.04 |
|
|
|
| chr21_-_19883182 | 1.04 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_-_33296077 | 1.04 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr2_+_49343278 | 1.02 |
ENSDART00000175147
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr10_-_8088063 | 1.01 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr15_-_23849212 | 0.99 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr14_+_20941526 | 0.97 |
ENSDART00000138551
|
smim19
|
small integral membrane protein 19 |
| chr7_+_13238684 | 0.94 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr5_-_53907908 | 0.93 |
ENSDART00000158069
|
ssna1
|
Sjogren syndrome nuclear autoantigen 1 |
| chr3_-_31487744 | 0.92 |
ENSDART00000124559
|
moto
|
minamoto |
| chr8_-_1833095 | 0.92 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr20_-_3292418 | 0.89 |
ENSDART00000099315
|
cebpz
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr23_+_44730111 | 0.88 |
ENSDART00000177271
|
trappc1
|
trafficking protein particle complex 1 |
| chr19_-_12473163 | 0.88 |
ENSDART00000165518
|
ptpn2b
|
protein tyrosine phosphatase, non-receptor type 2, b |
| chr22_-_510040 | 0.87 |
ENSDART00000140101
|
ccnd3
|
cyclin D3 |
| chr3_+_56882443 | 0.85 |
|
|
|
| chr21_-_19883409 | 0.84 |
ENSDART00000065670
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr8_-_16689736 | 0.82 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr24_+_25871962 | 0.82 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr25_-_34596670 | 0.81 |
ENSDART00000171659
|
zgc:162611
|
zgc:162611 |
| chr8_+_19945820 | 0.81 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr2_-_55584106 | 0.79 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr15_-_34550732 | 0.76 |
ENSDART00000139934
|
agmo
|
alkylglycerol monooxygenase |
| chr19_-_7522951 | 0.76 |
ENSDART00000003544
|
gabpb2a
|
GA binding protein transcription factor, beta subunit 2a |
| chr6_-_54425831 | 0.75 |
ENSDART00000017230
|
snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr19_+_7072033 | 0.71 |
ENSDART00000158758
ENSDART00000160482 |
kifc1
|
kinesin family member C1 |
| chr21_-_39616469 | 0.69 |
ENSDART00000026766
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr3_-_2041373 | 0.65 |
ENSDART00000146552
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr12_+_10078043 | 0.65 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr7_-_66470152 | 0.65 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr3_+_56882467 | 0.64 |
|
|
|
| chr22_+_2575708 | 0.63 |
|
|
|
| chr2_-_37419017 | 0.62 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr20_+_6545194 | 0.62 |
ENSDART00000159829
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr18_-_35861482 | 0.61 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr11_-_6870835 | 0.61 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr11_-_6442547 | 0.60 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr23_+_2104589 | 0.60 |
|
|
|
| chr22_-_21820673 | 0.60 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr25_-_5805538 | 0.59 |
|
|
|
| chr21_-_21477462 | 0.58 |
ENSDART00000031205
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr2_-_12852199 | 0.58 |
|
|
|
| chr21_-_14719281 | 0.58 |
ENSDART00000067004
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr11_-_6442490 | 0.57 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr23_-_77607 | 0.57 |
|
|
|
| chr19_+_40590165 | 0.56 |
ENSDART00000049147
|
rbm48
|
RNA binding motif protein 48 |
| chr14_+_11864388 | 0.56 |
ENSDART00000146521
|
rhogd
|
ras homolog gene family, member Gd |
| chr15_+_47207100 | 0.56 |
ENSDART00000154481
|
stard10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr23_+_39718769 | 0.55 |
ENSDART00000034690
|
otud3
|
OTU deubiquitinase 3 |
| chr18_-_17158425 | 0.55 |
ENSDART00000141873
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr23_-_25208472 | 0.54 |
ENSDART00000103989
ENSDART00000160278 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr7_-_66470118 | 0.54 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr7_+_23752492 | 0.53 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr20_-_3292250 | 0.52 |
ENSDART00000099315
|
cebpz
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr23_-_31986679 | 0.52 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr17_-_7377094 | 0.51 |
|
|
|
| chr8_-_53593850 | 0.51 |
|
|
|
| chr23_-_39879096 | 0.51 |
ENSDART00000159519
|
ENSDARG00000102199
|
ENSDARG00000102199 |
| chr4_-_25179188 | 0.51 |
ENSDART00000066929
|
taf3
|
TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated facto |
| chr3_+_30956719 | 0.50 |
ENSDART00000129564
|
si:dkey-66i24.8
|
si:dkey-66i24.8 |
| chr5_+_24378828 | 0.50 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr19_+_12487051 | 0.47 |
ENSDART00000052238
ENSDART00000013865 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr14_-_880729 | 0.46 |
ENSDART00000165211
|
rgs14a
|
regulator of G protein signaling 14a |
| chr5_-_66502280 | 0.45 |
ENSDART00000065264
|
cdca5
|
cell division cycle associated 5 |
| chr15_-_25431916 | 0.45 |
ENSDART00000047471
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr4_-_16893974 | 0.45 |
ENSDART00000124627
|
strap
|
serine/threonine kinase receptor associated protein |
| chr9_-_41352000 | 0.44 |
ENSDART00000059667
|
wdr75
|
WD repeat domain 75 |
| chr9_-_2522639 | 0.44 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr1_+_40428722 | 0.43 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr1_-_27395825 | 0.42 |
ENSDART00000170779
|
snapc3
|
small nuclear RNA activating complex, polypeptide 3 |
| chr20_-_40357240 | 0.42 |
|
|
|
| chr2_-_41722058 | 0.42 |
ENSDART00000022643
|
znf622
|
zinc finger protein 622 |
| chr4_-_11811833 | 0.41 |
ENSDART00000014153
|
cyb5r3
|
cytochrome b5 reductase 3 |
| chr13_-_49934345 | 0.40 |
ENSDART00000034541
|
gpatch11
|
G patch domain containing 11 |
| chr8_+_293186 | 0.40 |
ENSDART00000164099
ENSDART00000165472 |
snx2
|
sorting nexin 2 |
| chr2_+_9957535 | 0.39 |
ENSDART00000018524
|
pcyt1aa
|
phosphate cytidylyltransferase 1, choline, alpha a |
| chr8_-_26773755 | 0.38 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr18_-_19016801 | 0.37 |
ENSDART00000129776
|
vwa9
|
von Willebrand factor A domain containing 9 |
| chr2_-_43274603 | 0.36 |
ENSDART00000132346
ENSDART00000075347 |
lrrc6
|
leucine rich repeat containing 6 |
| chr11_+_17849528 | 0.35 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr21_-_2211701 | 0.34 |
ENSDART00000175686
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr20_-_51373292 | 0.33 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr22_-_29695242 | 0.32 |
|
|
|
| chr1_+_33471274 | 0.31 |
ENSDART00000046094
|
arl6
|
ADP-ribosylation factor-like 6 |
| chr23_-_31986571 | 0.30 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr25_+_386868 | 0.29 |
|
|
|
| chr24_+_25872051 | 0.27 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr9_-_35073289 | 0.25 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr14_+_33167301 | 0.25 |
ENSDART00000038128
|
mcts1
|
malignant T cell amplified sequence 1 |
| chr8_-_53593880 | 0.25 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr9_-_50106442 | 0.23 |
|
|
|
| chr8_-_38285034 | 0.23 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr7_-_13109764 | 0.22 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr17_+_12504341 | 0.22 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr18_-_50108685 | 0.22 |
|
|
|
| chr16_+_54350976 | 0.19 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr7_-_66469975 | 0.17 |
ENSDART00000021317
|
ctr9
|
CTR9 homolog, Paf1/RNA polymerase II complex component |
| chr7_-_39874079 | 0.17 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr23_-_31986482 | 0.16 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr13_-_35924447 | 0.16 |
ENSDART00000134955
|
lgmn
|
legumain |
| chr15_+_17138134 | 0.15 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr14_-_6831278 | 0.14 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr9_-_52950815 | 0.13 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr6_-_52348510 | 0.12 |
ENSDART00000142565
|
eif6
|
eukaryotic translation initiation factor 6 |
| chr24_+_26183653 | 0.11 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr11_+_29523717 | 0.11 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr23_-_45265411 | 0.09 |
|
|
|
| chr15_+_17094877 | 0.09 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr3_-_13311188 | 0.09 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr1_-_50438531 | 0.08 |
|
|
|
| chr25_+_35778740 | 0.08 |
ENSDART00000073404
|
zgc:114037
|
zgc:114037 |
| chr10_+_18919823 | 0.07 |
ENSDART00000138334
|
ppp2r2ab
|
protein phosphatase 2, regulatory subunit B, alpha b |
| chr12_-_29118423 | 0.07 |
ENSDART00000153175
|
si:ch211-214e3.5
|
si:ch211-214e3.5 |
| chr18_+_5616238 | 0.05 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr20_-_20455731 | 0.04 |
ENSDART00000170488
ENSDART00000047580 |
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr10_-_13158018 | 0.04 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr7_+_15065241 | 0.03 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr5_-_12060382 | 0.03 |
ENSDART00000133587
|
wsb2
|
WD repeat and SOCS box containing 2 |
| chr12_-_27497159 | 0.01 |
ENSDART00000066282
|
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.5 | 1.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.5 | 1.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 1.4 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.3 | 0.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 2.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.1 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.2 | 0.8 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.9 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 0.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 3.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 1.1 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 1.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0010984 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.4 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 2.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 0.2 | GO:0034553 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.0 | 1.0 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 1.3 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.5 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.6 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 1.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 1.0 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 0.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.6 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 1.0 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.7 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.1 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.0 | 0.5 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.0 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.2 | 1.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 1.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.1 | 1.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 3.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.0 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 0.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 3.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.9 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 3.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 0.9 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.2 | 1.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 1.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.2 | 2.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 2.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.6 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 2.0 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.4 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.4 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 1.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.7 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.9 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |