Project
DANIO-CODE
Navigation
Downloads
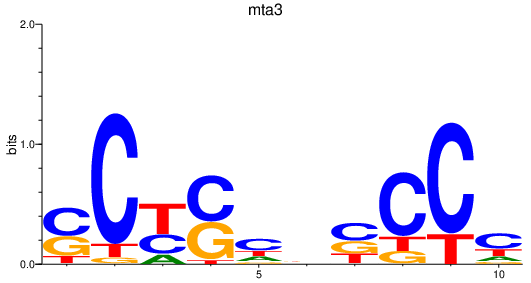
Results for mta3
Z-value: 0.40
Transcription factors associated with mta3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mta3
|
ENSDARG00000054903 | metastasis associated 1 family, member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mta3 | dr10_dc_chr12_+_25132396_25132577 | 0.53 | 3.6e-02 | Click! |
Activity profile of mta3 motif
Sorted Z-values of mta3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mta3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_33527870 | 0.97 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| KN149707v1_+_4014 | 0.86 |
ENSDART00000168516
|
cdo1
|
cysteine dioxygenase, type I |
| chr19_+_33267939 | 0.74 |
ENSDART00000176726
|
rpl30
|
ribosomal protein L30 |
| chr18_-_11216447 | 0.63 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr7_-_52283383 | 0.59 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr11_-_32461160 | 0.58 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr25_-_34638406 | 0.57 |
ENSDART00000123853
|
hist1h4l
|
histone 1, H4, like |
| chr19_+_42183946 | 0.53 |
ENSDART00000087096
|
tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr25_-_2485276 | 0.52 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr16_+_29109050 | 0.50 |
ENSDART00000122681
|
CR848841.1
|
Nestin |
| chr12_-_10372055 | 0.44 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr1_+_52308366 | 0.41 |
ENSDART00000108492
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr8_-_19872129 | 0.41 |
ENSDART00000129193
|
trabd2b
|
TraB domain containing 2B |
| chr20_-_54645287 | 0.35 |
ENSDART00000153389
|
yy1b
|
YY1 transcription factor b |
| chr5_+_68410884 | 0.34 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr21_+_44586326 | 0.29 |
|
|
|
| chr21_-_45595372 | 0.26 |
ENSDART00000163982
|
camlg
|
calcium modulating ligand |
| chr14_-_549816 | 0.26 |
ENSDART00000168951
|
fgf2
|
fibroblast growth factor 2 |
| chr3_-_5318206 | 0.25 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr21_+_25150600 | 0.23 |
ENSDART00000101147
|
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr1_+_52308299 | 0.20 |
ENSDART00000108492
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr13_-_33527029 | 0.20 |
|
|
|
| chr16_+_11138879 | 0.17 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr18_+_40364675 | 0.17 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr2_-_59013059 | 0.16 |
|
|
|
| chr13_-_4238224 | 0.16 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr5_+_65754237 | 0.15 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr5_-_21520111 | 0.14 |
|
|
|
| chr25_-_34601771 | 0.14 |
ENSDART00000073457
|
zgc:194989
|
zgc:194989 |
| chr6_-_12035327 | 0.12 |
|
|
|
| chr23_-_25866388 | 0.10 |
ENSDART00000138370
|
BX005414.1
|
ENSDARG00000094688 |
| chr4_-_13256905 | 0.10 |
ENSDART00000026593
ENSDART00000150577 |
grip1
|
glutamate receptor interacting protein 1 |
| chr8_+_27536121 | 0.09 |
ENSDART00000016696
|
rhocb
|
ras homolog family member Cb |
| chr20_+_54645315 | 0.08 |
|
|
|
| chr12_-_4351401 | 0.08 |
|
|
|
| chr5_+_30725475 | 0.08 |
|
|
|
| chr22_+_26840335 | 0.06 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr8_-_23250112 | 0.06 |
|
|
|
| chr25_-_2485217 | 0.04 |
ENSDART00000154889
ENSDART00000155027 |
BX957346.1
|
ENSDARG00000096850 |
| chr11_-_44280978 | 0.04 |
ENSDART00000099568
|
gpr137bb
|
G protein-coupled receptor 137Bb |
| chr14_-_2451650 | 0.04 |
ENSDART00000111748
|
pcdhb
|
protocadherin b |
| chr2_-_55971357 | 0.03 |
ENSDART00000154701
ENSDART00000154107 |
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr2_-_43318809 | 0.02 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr20_-_54645903 | 0.00 |
ENSDART00000038745
|
yy1b
|
YY1 transcription factor b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.4 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.3 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.1 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.9 | GO:0016702 | ferrous iron binding(GO:0008198) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |