Project
DANIO-CODE
Navigation
Downloads
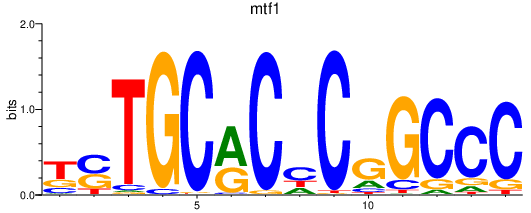
Results for mtf1
Z-value: 1.16
Transcription factors associated with mtf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mtf1
|
ENSDARG00000102898 | metal-regulatory transcription factor 1 |
Activity profile of mtf1 motif
Sorted Z-values of mtf1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mtf1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_471166 | 7.12 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr1_-_35196892 | 6.20 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr17_-_124685 | 4.49 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr8_+_31426328 | 4.01 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr22_-_37414940 | 3.77 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr3_+_23090476 | 2.70 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr23_+_23559246 | 2.60 |
ENSDART00000172214
|
agrn
|
agrin |
| chr1_+_44205158 | 2.59 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr22_-_11712371 | 2.59 |
|
|
|
| chr11_+_7518953 | 2.41 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr22_-_38621100 | 2.35 |
ENSDART00000164609
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr20_+_13998066 | 2.34 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr7_+_25052543 | 2.27 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr7_+_49442972 | 2.22 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr2_+_33474083 | 2.22 |
ENSDART00000056657
|
zgc:113531
|
zgc:113531 |
| chr4_-_28346021 | 2.14 |
ENSDART00000178149
|
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr14_-_36037883 | 2.00 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr17_-_53234493 | 1.96 |
|
|
|
| chr14_+_22889826 | 1.95 |
ENSDART00000170356
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr23_+_36079624 | 1.94 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr22_+_12327849 | 1.90 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr10_-_27035549 | 1.88 |
ENSDART00000146595
|
zgc:193742
|
zgc:193742 |
| chr17_+_37268262 | 1.88 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr1_+_49441608 | 1.83 |
ENSDART00000165973
ENSDART00000158988 ENSDART00000018469 |
npnt
|
nephronectin |
| chr20_-_158899 | 1.83 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr12_+_24536857 | 1.78 |
|
|
|
| chr12_-_17795448 | 1.78 |
|
|
|
| chr20_+_54464026 | 1.72 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr12_+_41016238 | 1.72 |
ENSDART00000170976
|
kif5bb
|
kinesin family member 5B, b |
| chr12_-_45714172 | 1.71 |
ENSDART00000149044
|
pax2b
|
paired box 2b |
| chr25_-_23428527 | 1.69 |
ENSDART00000062930
|
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr7_-_38590391 | 1.65 |
ENSDART00000037361
ENSDART00000173629 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr24_-_4418454 | 1.59 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr8_+_22334642 | 1.52 |
ENSDART00000146457
ENSDART00000142883 |
zgc:153631
|
zgc:153631 |
| chr8_+_999710 | 1.52 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr10_+_24476213 | 1.52 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr1_-_11591843 | 1.51 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr23_+_36079164 | 1.48 |
ENSDART00000103131
|
hoxc1a
|
homeo box C1a |
| chr1_-_54294303 | 1.40 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr5_-_55477243 | 1.38 |
|
|
|
| chr17_-_37040941 | 1.34 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr14_+_4810147 | 1.32 |
ENSDART00000114678
|
nanos1
|
nanos homolog 1 |
| chr2_+_46428154 | 1.31 |
ENSDART00000128457
|
ephb1
|
EPH receptor B1 |
| chr19_+_10912601 | 1.21 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr14_-_46407410 | 1.17 |
ENSDART00000173078
|
si:ch211-168f7.5
|
si:ch211-168f7.5 |
| chr16_+_11138924 | 1.16 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr20_+_54463948 | 1.15 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr20_-_46458839 | 1.04 |
ENSDART00000153087
|
bmf2
|
Bcl2 modifying factor 2 |
| chr1_-_11591684 | 1.02 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr5_+_8741824 | 1.02 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr23_+_45876483 | 1.01 |
ENSDART00000169521
ENSDART00000162915 |
dclk2b
|
doublecortin-like kinase 2b |
| chr16_+_11138879 | 1.01 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr6_+_7395817 | 0.98 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr5_+_37113743 | 0.95 |
|
|
|
| chr23_+_44137593 | 0.95 |
ENSDART00000148470
|
CU993818.1
|
ENSDARG00000095873 |
| chr24_-_3445036 | 0.92 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr24_-_38496534 | 0.90 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr24_-_3445303 | 0.89 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr8_-_51537858 | 0.89 |
ENSDART00000167394
|
fgfr1a
|
fibroblast growth factor receptor 1a |
| chr21_-_22441376 | 0.83 |
|
|
|
| chr11_+_7518748 | 0.74 |
ENSDART00000171813
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr20_-_1293519 | 0.73 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr23_+_23559198 | 0.73 |
ENSDART00000172214
|
agrn
|
agrin |
| chr17_+_25463178 | 0.72 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr3_+_23090404 | 0.72 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr23_-_18131105 | 0.70 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr1_+_8320586 | 0.66 |
ENSDART00000006211
|
prkcba
|
protein kinase C, beta a |
| chr9_+_50609926 | 0.66 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr8_+_8934762 | 0.64 |
ENSDART00000133137
|
bcap31
|
B-cell receptor-associated protein 31 |
| chr2_+_54911012 | 0.63 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr6_+_39056821 | 0.63 |
|
|
|
| chr23_+_45876447 | 0.61 |
ENSDART00000169521
ENSDART00000162915 |
dclk2b
|
doublecortin-like kinase 2b |
| chr17_+_37268209 | 0.61 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr4_+_47242957 | 0.60 |
|
|
|
| chr11_-_11487856 | 0.59 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr5_+_36014029 | 0.58 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr17_-_37040694 | 0.53 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr9_+_25109928 | 0.53 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_+_42027121 | 0.51 |
ENSDART00000138013
|
crlf1a
|
cytokine receptor-like factor 1a |
| chr9_-_53380782 | 0.46 |
ENSDART00000166906
|
trpm2
|
transient receptor potential cation channel, subfamily M, member 2 |
| chr24_-_25021505 | 0.45 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr13_+_40377712 | 0.44 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr20_+_571020 | 0.41 |
ENSDART00000152149
|
si:dkey-121j17.6
|
si:dkey-121j17.6 |
| chr7_-_69114789 | 0.41 |
|
|
|
| chr3_+_14151127 | 0.41 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr13_+_4040744 | 0.40 |
|
|
|
| chr7_-_38590218 | 0.39 |
ENSDART00000133491
|
phf21aa
|
PHD finger protein 21Aa |
| chr17_-_12231165 | 0.37 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr24_-_37989964 | 0.34 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr5_-_33611896 | 0.34 |
|
|
|
| chr9_+_41910191 | 0.34 |
|
|
|
| chr6_+_10214270 | 0.33 |
ENSDART00000090874
|
kcnh7
|
potassium channel, voltage gated eag related subfamily H, member 7 |
| chr13_+_4040802 | 0.32 |
|
|
|
| chr4_-_4698425 | 0.28 |
ENSDART00000093005
|
ENSDARG00000063578
|
ENSDARG00000063578 |
| chr23_-_18131209 | 0.26 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr7_+_27420387 | 0.25 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr7_+_64897785 | 0.25 |
ENSDART00000113602
ENSDART00000167822 |
ipo7
|
importin 7 |
| chr1_-_11591631 | 0.25 |
ENSDART00000142122
|
cplx2l
|
complexin 2, like |
| chr7_+_58866578 | 0.24 |
ENSDART00000167980
|
ostc
|
oligosaccharyltransferase complex subunit |
| chr19_+_43448344 | 0.22 |
ENSDART00000015632
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr10_+_21477806 | 0.20 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr11_-_3266080 | 0.20 |
ENSDART00000154314
|
prph
|
peripherin |
| chr3_+_38939974 | 0.18 |
|
|
|
| chr25_-_23428563 | 0.15 |
ENSDART00000062930
|
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr6_+_3120600 | 0.15 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr10_+_39142185 | 0.09 |
|
|
|
| chr7_+_20595694 | 0.09 |
ENSDART00000174012
|
gigyf1a
|
GRB10 interacting GYF protein 1a |
| chr15_-_3024027 | 0.08 |
|
|
|
| chr25_+_17217050 | 0.08 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr23_-_18131038 | 0.08 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr3_+_14151244 | 0.07 |
ENSDART00000162023
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr10_-_34122428 | 0.05 |
|
|
|
| chr3_-_18606063 | 0.02 |
ENSDART00000130029
|
zgc:113333
|
zgc:113333 |
| chr23_+_28472956 | 0.01 |
ENSDART00000135033
|
CT025585.1
|
ENSDARG00000093306 |
| chr1_-_54294382 | 0.01 |
ENSDART00000038330
|
khsrp
|
KH-type splicing regulatory protein |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 1.2 | 7.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 1.1 | 3.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.8 | 2.3 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.8 | 3.8 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.6 | 1.9 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.5 | 1.6 | GO:0060898 | radial glial cell differentiation(GO:0060019) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.5 | 1.8 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.4 | 1.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 3.4 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.3 | 0.9 | GO:0043589 | positive regulation of phospholipase C activity(GO:0010863) skin morphogenesis(GO:0043589) regulation of phospholipase C activity(GO:1900274) |
| 0.3 | 1.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 0.5 | GO:0050861 | regulation of antigen receptor-mediated signaling pathway(GO:0050854) regulation of B cell receptor signaling pathway(GO:0050855) positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 6.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.0 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 2.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.3 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.5 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 2.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.7 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 3.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 1.9 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 1.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 2.2 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 2.4 | GO:0072659 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 2.1 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 4.2 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.8 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.3 | 4.5 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.3 | 2.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 2.8 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.3 | GO:0060293 | germ plasm(GO:0060293) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 3.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.6 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.5 | 2.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 6.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.4 | 1.8 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.3 | 1.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 2.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 8.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 3.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 3.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.2 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 2.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 5.3 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 1.8 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 5.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 1.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 2.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 3.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.8 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |