Project
DANIO-CODE
Navigation
Downloads
Results for myb_mybl2a
Z-value: 0.70
Transcription factors associated with myb_mybl2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
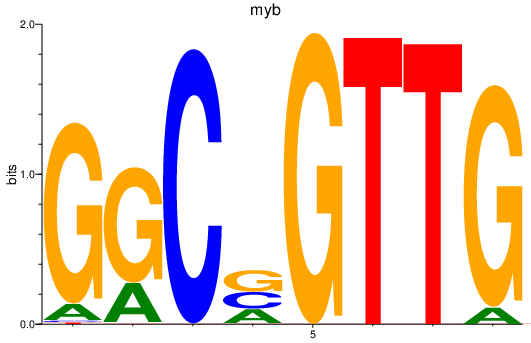
myb
|
ENSDARG00000053666 | v-myb avian myeloblastosis viral oncogene homolog |
|
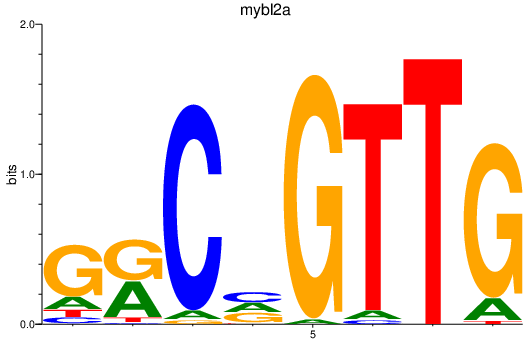
mybl2a
|
ENSDARG00000095241 | v-myb avian myeloblastosis viral oncogene homolog-like 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mybl2a | dr10_dc_chr23_+_4409342_4409553 | 0.70 | 2.6e-03 | Click! |
| myb | dr10_dc_chr23_+_31888903_31889033 | -0.61 | 1.3e-02 | Click! |
Activity profile of myb_mybl2a motif
Sorted Z-values of myb_mybl2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of myb_mybl2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_14042703 | 1.22 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr20_-_47576872 | 1.18 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr17_+_21797726 | 1.06 |
ENSDART00000079011
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr16_-_40508858 | 1.04 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr21_+_43183522 | 0.99 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr15_-_23786063 | 0.95 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr10_-_35313462 | 0.90 |
ENSDART00000139107
|
prr11
|
proline rich 11 |
| chr22_-_8144696 | 0.90 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr4_-_17289857 | 0.89 |
ENSDART00000178686
|
lrmp
|
lymphoid-restricted membrane protein |
| chr1_+_50893368 | 0.88 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr17_+_21797517 | 0.87 |
ENSDART00000079011
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr21_-_38670059 | 0.84 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr19_+_10742387 | 0.84 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr16_-_40508938 | 0.83 |
ENSDART00000032389
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr7_+_17695289 | 0.80 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr25_+_27301180 | 0.79 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr3_-_2041373 | 0.78 |
ENSDART00000146552
|
polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr8_-_53625018 | 0.78 |
|
|
|
| chr10_+_35313772 | 0.77 |
ENSDART00000028940
|
styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr24_+_21030141 | 0.76 |
ENSDART00000154940
|
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr16_-_31834830 | 0.74 |
|
|
|
| chr17_-_29253770 | 0.73 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr13_+_22973764 | 0.73 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr3_+_29338315 | 0.73 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr17_-_29253918 | 0.71 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr2_+_30497550 | 0.69 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr5_-_3323004 | 0.69 |
|
|
|
| chr9_-_30997261 | 0.68 |
ENSDART00000146300
|
klf5b
|
Kruppel-like factor 5b |
| chr3_-_36940649 | 0.67 |
ENSDART00000110716
ENSDART00000160233 |
tubg1
|
tubulin, gamma 1 |
| chr6_-_50686517 | 0.65 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr8_-_337744 | 0.64 |
|
|
|
| chr7_-_73620443 | 0.64 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr19_-_19452999 | 0.63 |
ENSDART00000163359
ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr14_-_25637853 | 0.63 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr23_+_4409342 | 0.62 |
ENSDART00000136287
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr19_+_10742887 | 0.62 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr7_-_15942989 | 0.61 |
ENSDART00000173739
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr15_-_643531 | 0.61 |
ENSDART00000102257
|
alox5b.3
|
arachidonate 5-lipoxygenase b, tandem duplicate 3 |
| chr20_+_33841199 | 0.61 |
ENSDART00000015000
|
henmt1
|
HEN1 methyltransferase homolog 1 (Arabidopsis) |
| chr25_+_29219700 | 0.60 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr16_-_31835007 | 0.60 |
|
|
|
| chr21_+_26954898 | 0.60 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr6_-_24292584 | 0.59 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr15_-_25158558 | 0.59 |
ENSDART00000142609
|
exo5
|
exonuclease 5 |
| chr19_+_10742944 | 0.58 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr12_-_24711074 | 0.58 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr22_-_12738593 | 0.57 |
ENSDART00000143574
|
rqcd1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr14_-_38942 | 0.57 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr6_+_40835073 | 0.57 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr6_+_4000356 | 0.55 |
ENSDART00000111817
|
trim25l
|
tripartite motif containing 25, like |
| chr10_+_11079874 | 0.54 |
ENSDART00000047954
|
ugcg
|
UDP-glucose ceramide glucosyltransferase |
| chr3_-_2047084 | 0.54 |
|
|
|
| chr10_+_21487458 | 0.53 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr7_+_6402162 | 0.53 |
|
|
|
| chr15_-_25158683 | 0.53 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr25_-_36691657 | 0.53 |
ENSDART00000153789
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr7_+_36281268 | 0.52 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr19_-_18664720 | 0.52 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr6_-_6818607 | 0.52 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr1_-_53220810 | 0.51 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr22_-_8144599 | 0.51 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr9_+_35141690 | 0.50 |
ENSDART00000140851
ENSDART00000077800 |
tfdp1a
|
transcription factor Dp-1, a |
| chr17_-_29254052 | 0.50 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr6_-_33931989 | 0.49 |
ENSDART00000141483
|
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr9_-_21677941 | 0.49 |
ENSDART00000121939
ENSDART00000080404 |
mphosph8
|
M-phase phosphoprotein 8 |
| chr2_+_30497691 | 0.49 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr8_-_14571519 | 0.48 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr11_+_16017857 | 0.47 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr5_+_56896961 | 0.47 |
|
|
|
| chr16_+_5625456 | 0.47 |
|
|
|
| chr15_-_23786239 | 0.46 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr19_+_10742594 | 0.46 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr10_+_21487218 | 0.46 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr6_-_40834531 | 0.46 |
ENSDART00000007783
|
eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr22_-_38321005 | 0.46 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr15_-_161052 | 0.46 |
|
|
|
| chr16_+_34038263 | 0.45 |
ENSDART00000134946
|
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr16_-_31834774 | 0.45 |
|
|
|
| chr11_-_13283709 | 0.45 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr15_+_17407036 | 0.45 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr16_-_31835083 | 0.45 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr7_-_71338273 | 0.44 |
|
|
|
| chr7_-_6209659 | 0.44 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr21_-_39521698 | 0.44 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr21_+_11943637 | 0.44 |
ENSDART00000141306
|
zgc:162344
|
zgc:162344 |
| chr14_+_30434569 | 0.43 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr16_+_48674071 | 0.43 |
ENSDART00000169738
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr19_-_33356861 | 0.43 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr22_-_25592743 | 0.43 |
ENSDART00000110638
ENSDART00000172092 |
si:ch211-12h2.8
|
si:ch211-12h2.8 |
| chr19_+_10742807 | 0.42 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr11_+_13118866 | 0.42 |
ENSDART00000123257
|
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr15_+_19948869 | 0.41 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr17_-_25630635 | 0.41 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr21_-_26379011 | 0.41 |
|
|
|
| chr17_+_49537268 | 0.41 |
ENSDART00000123641
|
zgc:113372
|
zgc:113372 |
| chr10_-_35488523 | 0.41 |
|
|
|
| chr14_-_47210912 | 0.41 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr16_-_31834945 | 0.41 |
|
|
|
| chr14_+_14850200 | 0.40 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr6_-_33886299 | 0.40 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr19_-_7606394 | 0.40 |
ENSDART00000092256
|
mrpl9
|
mitochondrial ribosomal protein L9 |
| chr15_+_6462943 | 0.40 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr19_+_10742344 | 0.39 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr6_+_33947023 | 0.39 |
ENSDART00000157397
|
orc1
|
origin recognition complex, subunit 1 |
| chr18_-_25784843 | 0.38 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr5_+_19950164 | 0.38 |
ENSDART00000008402
|
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr20_-_31840932 | 0.38 |
ENSDART00000137679
|
sash1a
|
SAM and SH3 domain containing 1a |
| chr8_-_14571436 | 0.38 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr17_-_33764220 | 0.38 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr6_-_7854902 | 0.37 |
|
|
|
| chr10_-_35304538 | 0.37 |
ENSDART00000087477
|
smg8
|
SMG8 nonsense mediated mRNA decay factor |
| chr15_-_24007354 | 0.37 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr17_-_29254184 | 0.37 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr13_+_1001437 | 0.37 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr7_-_15943107 | 0.37 |
ENSDART00000173492
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr25_+_27301039 | 0.37 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr21_+_11943609 | 0.37 |
ENSDART00000155426
ENSDART00000102463 |
zgc:162344
|
zgc:162344 |
| chr8_-_2557556 | 0.36 |
ENSDART00000140033
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr10_-_28230992 | 0.36 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr16_+_34038475 | 0.36 |
ENSDART00000130540
|
zdhhc18a
|
zinc finger, DHHC-type containing 18a |
| chr23_-_12310778 | 0.36 |
ENSDART00000131256
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr8_+_59148 | 0.36 |
ENSDART00000159739
ENSDART00000122378 |
cep120
|
centrosomal protein 120 |
| chr15_-_24007488 | 0.36 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr17_+_5680342 | 0.36 |
ENSDART00000156089
|
CU929322.1
|
ENSDARG00000096839 |
| chr7_-_30353095 | 0.36 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr24_-_6647047 | 0.35 |
|
|
|
| chr15_+_19948916 | 0.35 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr22_-_12738445 | 0.35 |
ENSDART00000047464
|
rqcd1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr12_-_43161273 | 0.35 |
|
|
|
| chr21_+_20350218 | 0.35 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr25_+_29219612 | 0.35 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr13_+_12538980 | 0.34 |
ENSDART00000010517
|
eif4eb
|
eukaryotic translation initiation factor 4eb |
| chr11_-_35894977 | 0.34 |
ENSDART00000138609
|
gpx1a
|
glutathione peroxidase 1a |
| chr15_-_24007305 | 0.34 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr18_+_39125694 | 0.34 |
ENSDART00000122377
|
bcl2l10
|
BCL2-like 10 (apoptosis facilitator) |
| chr17_-_49537093 | 0.33 |
ENSDART00000157331
ENSDART00000065917 ENSDART00000164715 |
zgc:171599
|
zgc:171599 |
| chr25_+_27301351 | 0.33 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr23_-_18105560 | 0.33 |
|
|
|
| chr6_+_28001359 | 0.33 |
ENSDART00000139367
|
amotl2a
|
angiomotin like 2a |
| chr11_+_44414503 | 0.33 |
|
|
|
| chr18_+_46153484 | 0.33 |
ENSDART00000015034
|
blvrb
|
biliverdin reductase B |
| chr16_+_38209634 | 0.33 |
ENSDART00000160332
|
pi4kb
|
phosphatidylinositol 4-kinase, catalytic, beta |
| chr21_+_19381959 | 0.33 |
ENSDART00000080110
|
amacr
|
alpha-methylacyl-CoA racemase |
| chr3_+_31585544 | 0.31 |
ENSDART00000135046
|
dcaf7
|
ddb1 and cul4 associated factor 7 |
| chr3_-_59690168 | 0.31 |
ENSDART00000035878
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr13_-_23626146 | 0.31 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr13_-_5128884 | 0.31 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr6_-_33946853 | 0.31 |
ENSDART00000057290
|
prpf38a
|
pre-mRNA processing factor 38A |
| chr1_+_57267909 | 0.31 |
ENSDART00000152640
|
BX469930.2
|
ENSDARG00000096615 |
| chr22_-_22234380 | 0.31 |
ENSDART00000020937
|
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr22_-_22234212 | 0.31 |
ENSDART00000113824
ENSDART00000169594 ENSDART00000158261 |
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr9_+_47348002 | 0.31 |
ENSDART00000176408
|
FP103056.1
|
ENSDARG00000108291 |
| chr14_-_16118942 | 0.31 |
|
|
|
| chr6_+_60117223 | 0.31 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr1_+_40894908 | 0.30 |
ENSDART00000111367
|
si:dkey-56e3.3
|
si:dkey-56e3.3 |
| chr5_-_35997345 | 0.30 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr2_-_8002471 | 0.30 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr19_+_33551956 | 0.30 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr2_+_35134340 | 0.30 |
ENSDART00000113609
|
rfwd2
|
ring finger and WD repeat domain 2 |
| chr6_-_10574082 | 0.30 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr17_-_33764256 | 0.30 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr24_+_21030422 | 0.30 |
ENSDART00000154940
|
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_+_22973830 | 0.30 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr22_+_10202801 | 0.30 |
ENSDART00000105900
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr20_+_41351373 | 0.29 |
ENSDART00000153067
|
BX510919.1
|
ENSDARG00000096764 |
| chr22_-_22139268 | 0.29 |
ENSDART00000101659
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr14_+_42109615 | 0.29 |
|
|
|
| chr6_-_17903824 | 0.29 |
ENSDART00000170369
|
nol11
|
nucleolar protein 11 |
| chr19_-_19883522 | 0.29 |
|
|
|
| chr15_+_19948426 | 0.29 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr3_+_33608669 | 0.29 |
|
|
|
| chr20_-_33272243 | 0.29 |
ENSDART00000178752
ENSDART00000047834 |
nbas
|
neuroblastoma amplified sequence |
| chr24_+_17001576 | 0.29 |
ENSDART00000149744
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr8_+_22268039 | 0.28 |
ENSDART00000075126
|
b3gnt7l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7, like |
| chr23_-_903314 | 0.28 |
ENSDART00000165102
|
rhoac
|
ras homolog gene family, member Ac |
| chr16_-_41787636 | 0.28 |
ENSDART00000121894
|
CR376740.2
|
ENSDARG00000097581 |
| chr8_-_16479425 | 0.28 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr15_+_19948680 | 0.28 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr2_-_24909432 | 0.28 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr16_-_39317068 | 0.28 |
ENSDART00000133642
|
gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr22_+_33180599 | 0.28 |
ENSDART00000004504
|
dag1
|
dystroglycan 1 |
| chr24_-_36413059 | 0.28 |
ENSDART00000062736
|
coasy
|
CoA synthase |
| chr5_-_60904215 | 0.28 |
ENSDART00000050912
|
pex12
|
peroxisomal biogenesis factor 12 |
| chr12_-_16385963 | 0.27 |
ENSDART00000152271
|
pcgf5b
|
polycomb group ring finger 5b |
| chr15_+_11772499 | 0.27 |
|
|
|
| chr3_+_52901875 | 0.27 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr18_-_16948109 | 0.27 |
ENSDART00000100117
|
znf143b
|
zinc finger protein 143b |
| chr1_-_15995931 | 0.27 |
ENSDART00000158166
|
frg1
|
FSHD region gene 1 |
| chr24_-_33894062 | 0.27 |
ENSDART00000079210
|
cdk5
|
cyclin-dependent kinase 5 |
| chr12_-_24710877 | 0.27 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr13_-_35782121 | 0.27 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr12_-_18446766 | 0.27 |
ENSDART00000039693
|
pgp
|
phosphoglycolate phosphatase |
| chr5_-_66070385 | 0.26 |
ENSDART00000032909
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr13_-_24695927 | 0.26 |
ENSDART00000087786
|
slka
|
STE20-like kinase a |
| chr16_-_52847564 | 0.26 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr6_-_32100075 | 0.26 |
ENSDART00000130627
|
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr3_-_86374 | 0.26 |
ENSDART00000138302
|
zgc:110249
|
zgc:110249 |
| chr21_+_20350334 | 0.26 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr24_+_14396536 | 0.26 |
ENSDART00000135646
|
ngdn
|
neuroguidin, EIF4E binding protein |
| chr6_+_40835289 | 0.26 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.3 | 1.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 0.9 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.3 | 0.8 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 3.3 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 0.6 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 0.6 | GO:0071887 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.2 | 2.3 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.2 | GO:0070922 | small RNA loading onto RISC(GO:0070922) |
| 0.2 | 0.6 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 1.1 | GO:0070601 | establishment of mitotic sister chromatid cohesion(GO:0034087) centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.7 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.1 | 0.7 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.6 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.6 | GO:0046476 | glucosylceramide metabolic process(GO:0006678) glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.4 | GO:0061511 | cell proliferation in forebrain(GO:0021846) centriole elongation(GO:0061511) |
| 0.1 | 0.4 | GO:1902633 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.1 | 0.3 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.5 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.5 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.3 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.1 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.8 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.4 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.7 | GO:0033292 | somatic muscle development(GO:0007525) T-tubule organization(GO:0033292) |
| 0.1 | 1.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.7 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.3 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.2 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) female pregnancy(GO:0007565) embryo implantation(GO:0007566) multi-multicellular organism process(GO:0044706) apelin receptor signaling pathway(GO:0060183) trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.3 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.3 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.2 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.6 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.5 | GO:0051923 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) sulfation(GO:0051923) |
| 0.0 | 2.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.3 | GO:0010522 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.0 | 0.3 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.1 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.5 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 1.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.9 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0071922 | regulation of sister chromatid cohesion(GO:0007063) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.6 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.7 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.0 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.3 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.3 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 0.6 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.6 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.0 | 1.0 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 1.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.6 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.6 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.3 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.3 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.4 | 1.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.3 | 1.0 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 0.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 1.1 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 0.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.2 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 0.5 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.2 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.4 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 0.4 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 1.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.1 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.2 | GO:0030882 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.2 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.4 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 1.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.0 | 0.2 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.1 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.0 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.9 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 2.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |