Project
DANIO-CODE
Navigation
Downloads
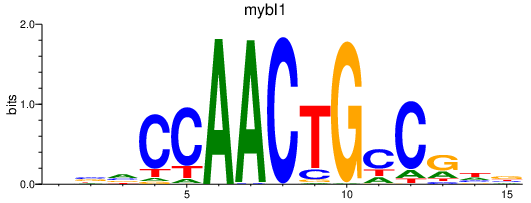
Results for mybl1
Z-value: 1.46
Transcription factors associated with mybl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mybl1
|
ENSDARG00000030999 | v-myb avian myeloblastosis viral oncogene homolog-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mybl1 | dr10_dc_chr24_+_23646663_23646747 | -0.53 | 3.6e-02 | Click! |
Activity profile of mybl1 motif
Sorted Z-values of mybl1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mybl1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25740782 | 4.69 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr14_-_35552296 | 3.77 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr14_+_30434569 | 3.63 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr22_-_8144696 | 3.37 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr23_+_4409342 | 3.34 |
ENSDART00000136287
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr22_-_28828375 | 3.22 |
ENSDART00000104880
ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr13_-_42548129 | 3.01 |
ENSDART00000169083
|
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr21_+_28710341 | 3.01 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr1_+_50893368 | 2.99 |
ENSDART00000152789
|
etaa1
|
ETAA1, ATR kinase activator |
| chr18_+_17545564 | 2.83 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr22_+_31073990 | 2.71 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr20_+_30476065 | 2.65 |
ENSDART00000062441
|
rnaseh1
|
ribonuclease H1 |
| chr14_+_30434751 | 2.62 |
ENSDART00000139975
|
atl3
|
atlastin 3 |
| chr5_+_56896961 | 2.58 |
|
|
|
| chr22_-_8144599 | 2.56 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr1_+_45784486 | 2.47 |
|
|
|
| chr22_-_7976467 | 2.35 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr3_+_22914624 | 2.26 |
ENSDART00000157090
ENSDART00000103858 |
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr3_-_3646681 | 2.16 |
ENSDART00000165648
ENSDART00000046454 ENSDART00000115282 ENSDART00000164463 |
ENSDARG00000076771
|
ENSDARG00000076771 |
| chr18_-_16947996 | 2.14 |
ENSDART00000100117
|
znf143b
|
zinc finger protein 143b |
| chr18_-_16948109 | 2.08 |
ENSDART00000100117
|
znf143b
|
zinc finger protein 143b |
| chr6_-_46740359 | 2.05 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr7_-_15942989 | 1.94 |
ENSDART00000173739
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr11_-_22210506 | 1.91 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr3_+_22914688 | 1.88 |
ENSDART00000156472
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr20_+_2100836 | 1.85 |
|
|
|
| chr2_-_24909546 | 1.84 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr7_-_15943107 | 1.80 |
ENSDART00000173492
|
btr04
|
bloodthirsty-related gene family, member 4 |
| chr21_-_26655258 | 1.79 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr15_-_24007305 | 1.74 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr21_+_28710269 | 1.70 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr21_+_28710424 | 1.63 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr1_-_42915971 | 1.58 |
|
|
|
| chr2_-_24909432 | 1.54 |
ENSDART00000170283
|
crsp7
|
cofactor required for Sp1 transcriptional activation, subunit 7 |
| chr6_-_22311576 | 1.44 |
|
|
|
| chr6_-_6818607 | 1.44 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr12_+_31523147 | 1.43 |
ENSDART00000153129
|
dnmbp
|
dynamin binding protein |
| chr1_+_35253862 | 1.40 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr3_+_14421976 | 1.39 |
ENSDART00000171731
|
znf653
|
zinc finger protein 653 |
| chr20_+_30475949 | 1.34 |
ENSDART00000148242
|
rnaseh1
|
ribonuclease H1 |
| chr6_-_9394499 | 1.34 |
ENSDART00000013066
|
ercc3
|
excision repair cross-complementation group 3 |
| chr17_-_43632707 | 1.34 |
ENSDART00000136431
|
rtkn2a
|
rhotekin 2a |
| chr20_+_5022501 | 1.31 |
ENSDART00000162678
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr10_-_13549869 | 1.30 |
|
|
|
| chr2_+_30497550 | 1.29 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr13_+_15571152 | 1.29 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr14_+_6228658 | 1.23 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr1_+_44248486 | 1.21 |
ENSDART00000067621
|
vps37c
|
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
| chr21_-_19879125 | 1.18 |
ENSDART00000164317
|
eri1
|
exoribonuclease 1 |
| chr13_-_37043725 | 1.13 |
|
|
|
| chr2_+_30497691 | 1.11 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr14_+_7596207 | 1.10 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr5_+_56896748 | 1.09 |
|
|
|
| chr24_-_20812363 | 1.07 |
ENSDART00000142080
|
kpna1
|
karyopherin alpha 1 (importin alpha 5) |
| chr20_-_47576872 | 1.07 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr12_+_31523411 | 1.06 |
ENSDART00000153129
|
dnmbp
|
dynamin binding protein |
| chr11_+_25443646 | 1.05 |
ENSDART00000121478
|
otud5b
|
OTU deubiquitinase 5b |
| chr23_-_539640 | 1.03 |
ENSDART00000128788
|
mtg2
|
mitochondrial ribosome-associated GTPase 2 |
| chr20_+_7337986 | 1.03 |
ENSDART00000140222
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr24_+_14095601 | 1.01 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr5_-_39440181 | 1.00 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr23_+_33792005 | 0.96 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr23_+_40070965 | 0.96 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr16_-_31386496 | 0.95 |
|
|
|
| chr6_-_7854902 | 0.95 |
|
|
|
| chr7_-_24778134 | 0.94 |
ENSDART00000135179
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr3_-_6833379 | 0.93 |
ENSDART00000123724
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr15_-_24007488 | 0.92 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr8_+_44448437 | 0.88 |
ENSDART00000143807
|
mhc1laa
|
major histocompatibility complex class I LAA |
| chr20_+_31375136 | 0.86 |
|
|
|
| chr12_+_28685081 | 0.83 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr14_+_35552581 | 0.80 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr22_-_28828300 | 0.77 |
ENSDART00000104880
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr17_+_19461192 | 0.77 |
ENSDART00000024194
|
kif11
|
kinesin family member 11 |
| chr2_+_20655215 | 0.75 |
|
|
|
| chr6_-_17903824 | 0.72 |
ENSDART00000170369
|
nol11
|
nucleolar protein 11 |
| chr23_+_44937093 | 0.70 |
|
|
|
| chr15_-_24007354 | 0.68 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr20_+_31375056 | 0.67 |
|
|
|
| chr21_+_28710522 | 0.66 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr3_-_14421675 | 0.63 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr23_-_36469779 | 0.57 |
ENSDART00000168246
|
spryd3
|
SPRY domain containing 3 |
| chr17_-_1949701 | 0.54 |
ENSDART00000013093
ENSDART00000168231 ENSDART00000166833 |
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr22_-_20695416 | 0.53 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr13_+_31390033 | 0.53 |
ENSDART00000086018
|
pcnxl4
|
pecanex-like 4 (Drosophila) |
| chr20_+_5022399 | 0.50 |
ENSDART00000053883
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr20_-_26517489 | 0.48 |
|
|
|
| chr7_+_22538844 | 0.44 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr5_+_31382951 | 0.44 |
ENSDART00000007458
|
ppp6c
|
protein phosphatase 6, catalytic subunit |
| chr13_-_42547873 | 0.39 |
ENSDART00000169083
|
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr16_-_20044830 | 0.38 |
|
|
|
| chr7_+_51541162 | 0.36 |
ENSDART00000174120
|
si:ch211-122f10.4
|
si:ch211-122f10.4 |
| chr19_-_33356861 | 0.33 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr20_+_31372117 | 0.29 |
|
|
|
| chr3_-_14421728 | 0.29 |
ENSDART00000137197
|
swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr18_-_5619309 | 0.26 |
ENSDART00000177705
|
CABZ01081638.1
|
ENSDARG00000107008 |
| chr9_-_35003703 | 0.26 |
|
|
|
| chr7_-_31650421 | 0.24 |
ENSDART00000146720
|
lin7c
|
lin-7 homolog C (C. elegans) |
| chr14_+_46621228 | 0.23 |
ENSDART00000173248
|
si:ch211-276i12.11
|
si:ch211-276i12.11 |
| chr12_+_31523317 | 0.20 |
ENSDART00000153129
|
dnmbp
|
dynamin binding protein |
| chr24_+_17001271 | 0.18 |
ENSDART00000149744
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr24_+_14070379 | 0.17 |
ENSDART00000004664
|
tram1
|
translocation associated membrane protein 1 |
| chr5_+_40910195 | 0.16 |
ENSDART00000134317
|
vcp
|
valosin containing protein |
| chr24_-_30924897 | 0.15 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr1_+_35253821 | 0.14 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr5_-_39440105 | 0.07 |
ENSDART00000157755
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr16_+_24046933 | 0.07 |
|
|
|
| chr24_+_17001576 | 0.04 |
ENSDART00000149744
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr23_-_18131038 | 0.00 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.0 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.5 | 4.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.5 | 3.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.4 | 4.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.4 | 2.8 | GO:0010574 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.3 | 1.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.8 | GO:0042823 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 1.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 4.7 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.2 | 0.7 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.2 | 0.9 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 6.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.4 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.3 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.8 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.1 | GO:0098926 | postsynaptic signal transduction(GO:0098926) |
| 0.1 | 1.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 1.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 1.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.5 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.2 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.2 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.2 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0097196 | Shu complex(GO:0097196) |
| 0.3 | 3.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 3.0 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 4.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 2.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 1.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.3 | 1.3 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.2 | 3.8 | GO:0015144 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.2 | 1.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 4.1 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.5 | GO:0042979 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.1 | 1.0 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 1.1 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.3 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 2.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 4.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 8.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.7 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 4.8 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.2 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |