Project
DANIO-CODE
Navigation
Downloads
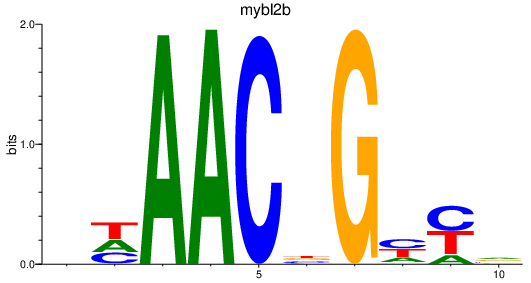
Results for mybl2b
Z-value: 2.61
Transcription factors associated with mybl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mybl2b
|
ENSDARG00000032264 | v-myb avian myeloblastosis viral oncogene homolog-like 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mybl2b | dr10_dc_chr11_+_1524965_1525123 | -0.56 | 2.4e-02 | Click! |
Activity profile of mybl2b motif
Sorted Z-values of mybl2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mybl2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_21495522 | 9.96 |
ENSDART00000105925
|
gtf3ab
|
general transcription factor IIIA, b |
| chr4_-_20456825 | 8.20 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr7_+_17695173 | 7.91 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr24_-_9857510 | 7.02 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr11_+_29299382 | 6.54 |
|
|
|
| chr22_-_28828375 | 6.45 |
ENSDART00000104880
ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr10_+_32107014 | 6.40 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr7_+_58448953 | 5.96 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr10_-_26240772 | 5.87 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr5_-_66145078 | 5.20 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr23_+_19663746 | 4.53 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr22_-_8144696 | 4.52 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr24_-_9865837 | 4.46 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr5_+_52199662 | 4.31 |
|
|
|
| chr3_+_16880947 | 4.29 |
ENSDART00000080854
|
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr11_+_29295032 | 4.19 |
ENSDART00000112721
|
BX548000.1
|
ENSDARG00000079546 |
| chr6_-_31196004 | 4.00 |
|
|
|
| chr20_+_791374 | 3.97 |
ENSDART00000137135
|
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr16_-_55210909 | 3.82 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr1_-_53052062 | 3.74 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr7_+_31154984 | 3.71 |
ENSDART00000138456
|
eif3jb
|
eukaryotic translation initiation factor 3, subunit Jb |
| chr13_+_22973764 | 3.69 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr23_-_3568408 | 3.65 |
ENSDART00000019667
|
rnf114
|
ring finger protein 114 |
| chr14_+_30434569 | 3.63 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr8_-_30732816 | 3.58 |
ENSDART00000098986
|
gucd1
|
guanylyl cyclase domain containing 1 |
| chr10_+_16543480 | 3.57 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr15_+_6462447 | 3.50 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr22_-_8144599 | 3.46 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr15_-_24007305 | 3.40 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr13_+_11417882 | 3.37 |
ENSDART00000034935
|
desi2
|
desumoylating isopeptidase 2 |
| chr7_+_35163845 | 3.33 |
ENSDART00000173733
|
BX294178.2
|
ENSDARG00000104955 |
| chr24_+_21495482 | 3.25 |
ENSDART00000105925
|
gtf3ab
|
general transcription factor IIIA, b |
| chr14_-_47387481 | 3.23 |
ENSDART00000031498
|
ccna2
|
cyclin A2 |
| chr7_+_26730804 | 3.22 |
|
|
|
| chr1_+_29919378 | 3.20 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr23_+_30803518 | 3.20 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr5_+_36066758 | 3.13 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr16_-_11350009 | 3.08 |
ENSDART00000134752
|
znf526
|
zinc finger protein 526 |
| chr23_+_30803887 | 3.05 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr5_+_67157818 | 3.03 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr8_-_44247277 | 2.98 |
|
|
|
| chr17_+_51655501 | 2.97 |
ENSDART00000149807
|
odc1
|
ornithine decarboxylase 1 |
| chr1_+_49170632 | 2.97 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr13_+_45387478 | 2.96 |
ENSDART00000019113
|
tmem57b
|
transmembrane protein 57b |
| chr5_+_36066651 | 2.96 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr8_-_14571519 | 2.96 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr4_-_1496779 | 2.95 |
ENSDART00000166360
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr14_-_23798158 | 2.94 |
ENSDART00000172747
|
si:ch211-277c7.7
|
si:ch211-277c7.7 |
| chr24_+_25872005 | 2.94 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr13_+_35799681 | 2.93 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr25_+_29873412 | 2.91 |
ENSDART00000067056
|
ticrr
|
TopBP1-interacting, checkpoint, and replication regulator |
| chr15_-_28636418 | 2.90 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr23_+_4409342 | 2.83 |
ENSDART00000136287
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr6_-_43297126 | 2.83 |
ENSDART00000154170
|
frmd4ba
|
FERM domain containing 4Ba |
| chr23_-_33811895 | 2.80 |
ENSDART00000131680
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr7_+_24119777 | 2.76 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr13_+_45387708 | 2.75 |
ENSDART00000074567
|
tmem57b
|
transmembrane protein 57b |
| chr25_+_27836260 | 2.75 |
ENSDART00000149373
|
aass
|
aminoadipate-semialdehyde synthase |
| chr17_-_29706238 | 2.74 |
ENSDART00000153865
|
CT027582.2
|
ENSDARG00000097817 |
| chr4_-_20456437 | 2.70 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr2_+_9854294 | 2.65 |
|
|
|
| chr1_-_50395003 | 2.64 |
ENSDART00000035150
|
spast
|
spastin |
| chr18_+_47305642 | 2.64 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr13_+_15571152 | 2.61 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr24_+_28855974 | 2.61 |
ENSDART00000017427
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr13_+_25234411 | 2.61 |
ENSDART00000027428
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr6_+_60117223 | 2.56 |
ENSDART00000008243
|
prelid3b
|
PRELI domain containing 3 |
| chr5_+_58564789 | 2.55 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr16_+_38400861 | 2.54 |
ENSDART00000133752
|
elp6
|
elongator acetyltransferase complex subunit 6 |
| chr3_+_12087549 | 2.53 |
|
|
|
| chr11_+_19440815 | 2.51 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr7_+_24119940 | 2.48 |
ENSDART00000108753
|
poln
|
polymerase (DNA directed) nu |
| chr6_+_43452686 | 2.47 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr20_+_791328 | 2.47 |
ENSDART00000137135
|
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr18_+_27457678 | 2.44 |
ENSDART00000014726
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr15_-_30247084 | 2.43 |
ENSDART00000154069
|
BX322555.4
|
ENSDARG00000097908 |
| chr1_+_31319012 | 2.43 |
ENSDART00000146602
|
sts
|
steroid sulfatase (microsomal), isozyme S |
| chr25_+_35790000 | 2.42 |
ENSDART00000152195
|
si:ch211-113a14.18
|
si:ch211-113a14.18 |
| chr24_-_36413059 | 2.41 |
ENSDART00000062736
|
coasy
|
CoA synthase |
| chr10_+_19059739 | 2.41 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr4_+_9010972 | 2.36 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr5_+_58564645 | 2.35 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 (IL11 inducible) |
| chr2_+_15379717 | 2.34 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr25_+_17829003 | 2.33 |
ENSDART00000067305
|
zgc:103499
|
zgc:103499 |
| chr8_+_19589519 | 2.31 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr5_-_8206388 | 2.30 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr23_-_24299965 | 2.30 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr17_+_23709676 | 2.30 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr5_+_52199767 | 2.28 |
|
|
|
| chr20_+_52968725 | 2.28 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr22_-_11799717 | 2.28 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr7_+_31155071 | 2.27 |
ENSDART00000138456
|
eif3jb
|
eukaryotic translation initiation factor 3, subunit Jb |
| chr19_+_29750506 | 2.26 |
ENSDART00000164432
|
ldlrap1a
|
low density lipoprotein receptor adaptor protein 1a |
| chr16_-_39317068 | 2.25 |
ENSDART00000133642
|
gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr20_+_44684340 | 2.25 |
ENSDART00000149775
|
atad2b
|
ATPase family, AAA domain containing 2B |
| chr17_-_12095843 | 2.24 |
ENSDART00000155545
|
ahctf1
|
AT hook containing transcription factor 1 |
| chr2_-_47827141 | 2.24 |
ENSDART00000056882
|
cul3a
|
cullin 3a |
| chr19_+_24741351 | 2.23 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr3_-_36940649 | 2.21 |
ENSDART00000110716
ENSDART00000160233 |
tubg1
|
tubulin, gamma 1 |
| chr7_+_68964813 | 2.21 |
ENSDART00000166258
|
marveld3
|
MARVEL domain containing 3 |
| chr17_+_34238753 | 2.20 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr15_+_25503446 | 2.19 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr8_+_59148 | 2.18 |
ENSDART00000159739
ENSDART00000122378 |
cep120
|
centrosomal protein 120 |
| chr19_-_46500888 | 2.15 |
ENSDART00000178772
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr10_+_16543577 | 2.14 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr7_-_38443332 | 2.11 |
ENSDART00000047220
|
fbxo3
|
F-box protein 3 |
| chr14_+_31189527 | 2.11 |
ENSDART00000053026
|
fam122b
|
family with sequence similarity 122B |
| chr17_-_43725091 | 2.11 |
|
|
|
| chr24_-_6304289 | 2.10 |
ENSDART00000140212
|
zgc:174877
|
zgc:174877 |
| chr10_+_21487458 | 2.10 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr5_+_36067010 | 2.09 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr15_-_24007488 | 2.06 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr22_-_7976467 | 2.05 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr21_+_7570420 | 2.04 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr3_+_13780104 | 2.04 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr18_-_510340 | 2.02 |
ENSDART00000164374
|
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr12_+_47828020 | 1.99 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr13_-_3805904 | 1.97 |
ENSDART00000017052
|
ncoa4
|
nuclear receptor coactivator 4 |
| chr4_-_10770022 | 1.97 |
|
|
|
| chr25_-_13394261 | 1.97 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr6_+_28001359 | 1.95 |
ENSDART00000139367
|
amotl2a
|
angiomotin like 2a |
| chr3_-_6833379 | 1.95 |
ENSDART00000123724
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr16_+_28857531 | 1.95 |
ENSDART00000161525
|
zgc:171704
|
zgc:171704 |
| chr5_+_6005049 | 1.94 |
|
|
|
| chr15_+_28477741 | 1.94 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr19_+_16159980 | 1.94 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr15_-_161052 | 1.93 |
|
|
|
| chr15_-_23588004 | 1.92 |
ENSDART00000078396
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr22_-_28828300 | 1.92 |
ENSDART00000104880
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr17_-_51729568 | 1.91 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr19_-_24551474 | 1.91 |
ENSDART00000104087
|
thap7
|
THAP domain containing 7 |
| chr9_-_35824470 | 1.90 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr16_-_4940519 | 1.90 |
ENSDART00000149421
|
rpa2
|
replication protein A2 |
| chr11_+_6872272 | 1.90 |
ENSDART00000075998
|
klhl26
|
kelch-like family member 26 |
| chr14_+_15788442 | 1.88 |
ENSDART00000159181
|
immt
|
inner membrane protein, mitochondrial (mitofilin) |
| chr7_-_73625494 | 1.87 |
ENSDART00000041385
|
zgc:163061
|
zgc:163061 |
| chr15_+_6462943 | 1.86 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr17_+_23463668 | 1.85 |
|
|
|
| chr14_+_30434751 | 1.84 |
ENSDART00000139975
|
atl3
|
atlastin 3 |
| chr9_-_18807868 | 1.83 |
|
|
|
| chr11_-_12174576 | 1.82 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr21_-_21142020 | 1.81 |
ENSDART00000079678
|
ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr20_+_791476 | 1.80 |
ENSDART00000137135
|
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr4_-_20456517 | 1.80 |
ENSDART00000003621
ENSDART00000132356 |
sinup
|
siaz-interacting nuclear protein |
| chr22_-_7887341 | 1.78 |
ENSDART00000097198
|
sc:d217
|
sc:d217 |
| chr24_-_36412892 | 1.77 |
ENSDART00000148868
|
coasy
|
CoA synthase |
| chr19_-_25843075 | 1.77 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr8_-_41511380 | 1.76 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr8_+_23360660 | 1.76 |
ENSDART00000142783
|
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr1_-_49307663 | 1.74 |
|
|
|
| chr10_+_21487218 | 1.74 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr5_-_62389958 | 1.71 |
|
|
|
| chr3_+_53097692 | 1.70 |
ENSDART00000082715
|
camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr15_-_23587867 | 1.69 |
ENSDART00000078396
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr3_-_27470798 | 1.69 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr15_+_31709330 | 1.69 |
ENSDART00000159634
|
b3glcta
|
beta 3-glucosyltransferase a |
| chr15_-_24007354 | 1.68 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr2_+_31959127 | 1.67 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr5_-_68751287 | 1.67 |
ENSDART00000112692
|
ENSDARG00000077155
|
ENSDARG00000077155 |
| chr8_-_14571436 | 1.67 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr5_+_4092360 | 1.66 |
ENSDART00000158826
|
CABZ01058650.1
|
ENSDARG00000101063 |
| chr3_+_16880842 | 1.65 |
ENSDART00000080854
|
stat3
|
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr2_+_35750334 | 1.64 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr10_-_25059072 | 1.64 |
ENSDART00000040733
|
cd209
|
CD209 molecule |
| chr7_-_73625414 | 1.63 |
ENSDART00000041385
|
zgc:163061
|
zgc:163061 |
| chr17_-_12095723 | 1.63 |
ENSDART00000115321
|
ahctf1
|
AT hook containing transcription factor 1 |
| chr19_-_12158958 | 1.63 |
ENSDART00000024193
|
rnf19a
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr25_-_13307171 | 1.63 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr17_+_26785230 | 1.62 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr7_+_58448909 | 1.62 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr3_-_45018353 | 1.62 |
ENSDART00000166146
|
dnajb1a
|
DnaJ (Hsp40) homolog, subfamily B, member 1a |
| chr16_-_11350199 | 1.62 |
ENSDART00000134752
|
znf526
|
zinc finger protein 526 |
| chr17_-_22553428 | 1.62 |
ENSDART00000079390
|
exo1
|
exonuclease 1 |
| chr24_-_5904097 | 1.60 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr8_+_23126036 | 1.60 |
ENSDART00000141175
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr7_+_29930406 | 1.59 |
ENSDART00000075588
|
wdr76
|
WD repeat domain 76 |
| chr19_-_35863647 | 1.59 |
ENSDART00000179357
|
anln
|
anillin, actin binding protein |
| chr18_+_1018614 | 1.57 |
ENSDART00000161206
|
pkma
|
pyruvate kinase, muscle, a |
| chr19_+_10640291 | 1.57 |
ENSDART00000151801
|
usf2
|
upstream transcription factor 2, c-fos interacting |
| chr3_+_45375580 | 1.56 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr1_-_44227583 | 1.55 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr1_-_39384698 | 1.54 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr10_+_16544000 | 1.53 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr8_-_49739908 | 1.53 |
ENSDART00000138810
|
gkap1
|
G kinase anchoring protein 1 |
| chr8_-_31407839 | 1.52 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr6_-_6818607 | 1.51 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr21_-_40556480 | 1.51 |
ENSDART00000168850
|
taok1b
|
TAO kinase 1b |
| chr6_-_33886299 | 1.51 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr10_+_40405689 | 1.51 |
|
|
|
| chr8_-_41511321 | 1.50 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr7_-_69114652 | 1.50 |
|
|
|
| chr15_-_28636615 | 1.50 |
ENSDART00000127845
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr16_-_41854586 | 1.49 |
ENSDART00000134718
|
psmc4
|
proteasome 26S subunit, ATPase 4 |
| chr10_-_26240909 | 1.48 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr17_+_13292569 | 1.46 |
ENSDART00000177104
|
BX005308.2
|
ENSDARG00000108498 |
| chr25_-_13562558 | 1.46 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr6_+_19933322 | 1.45 |
ENSDART00000143378
|
thumpd3
|
THUMP domain containing 3 |
| chr18_-_16927749 | 1.45 |
ENSDART00000100105
ENSDART00000163400 |
swap70b
|
SWAP switching B-cell complex 70b subunit 70b |
| chr22_-_6490288 | 1.45 |
ENSDART00000149676
|
CR352226.7
|
ENSDARG00000095952 |
| chr11_-_25223594 | 1.45 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 5.9 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 1.2 | 7.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 1.0 | 4.1 | GO:0051977 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 1.0 | 2.9 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 1.0 | 3.9 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 1.0 | 3.8 | GO:0010482 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.9 | 4.4 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.9 | 3.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.7 | 2.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.7 | 2.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.7 | 2.8 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.7 | 2.6 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.7 | 2.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.6 | 12.7 | GO:0001840 | neural plate development(GO:0001840) |
| 0.6 | 6.0 | GO:0097324 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.6 | 3.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.6 | 2.9 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.5 | 4.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 3.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.5 | 2.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.5 | 3.0 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.5 | 1.5 | GO:0014015 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.5 | 3.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.5 | 1.4 | GO:0097704 | response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.4 | 2.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 2.6 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.4 | 2.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 1.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.4 | 0.8 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.4 | 1.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.4 | 1.9 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.4 | 1.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.4 | 3.3 | GO:0070193 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.4 | 2.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 1.7 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.3 | 1.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.3 | 1.6 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.3 | 2.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 7.4 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.3 | 0.6 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.3 | 2.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 1.8 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 1.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.3 | 4.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.3 | 2.0 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.3 | 4.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.3 | 6.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.3 | 1.1 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.3 | 1.6 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.3 | 3.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 1.9 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 1.6 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 3.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 1.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.8 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.2 | 1.3 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 1.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.9 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.2 | 1.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 1.0 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 2.9 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.2 | 4.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.2 | 5.9 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 1.7 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.2 | 1.0 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) |
| 0.2 | 5.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 0.5 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.2 | 0.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.4 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 2.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.0 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 2.0 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 0.5 | GO:0044878 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 4.3 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.4 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 2.2 | GO:0010632 | regulation of epithelial cell migration(GO:0010632) |
| 0.1 | 3.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 0.7 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.1 | 3.9 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 4.4 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.2 | GO:0051646 | mitochondrion localization(GO:0051646) |
| 0.1 | 2.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 2.1 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 4.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.1 | 4.9 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 1.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 1.6 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 1.2 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 4.6 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.1 | 1.2 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 3.5 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 1.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 1.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.9 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 9.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.3 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 1.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.3 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.7 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 1.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 4.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.0 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 3.6 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 1.3 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.7 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 1.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 1.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.6 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.8 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 1.6 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.7 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 2.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0070663 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 0.6 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 2.9 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 2.5 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 1.6 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 8.0 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 0.6 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 1.0 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 1.6 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 1.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 1.1 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.5 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.2 | GO:0000800 | lateral element(GO:0000800) |
| 1.5 | 7.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 1.1 | 3.3 | GO:0000801 | central element(GO:0000801) |
| 1.1 | 3.2 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.5 | 2.6 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.5 | 2.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.4 | 1.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.4 | 3.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 2.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.4 | 1.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.4 | 2.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.4 | 2.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 2.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 2.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 2.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 3.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.3 | 1.6 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 2.5 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.2 | GO:0017054 | negative cofactor 2 complex(GO:0017054) |
| 0.3 | 0.9 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.3 | 2.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 1.7 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 1.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 1.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.2 | 1.9 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 4.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.2 | 1.0 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 1.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 3.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 9.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 4.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 5.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 7.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.8 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.1 | 7.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.8 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 2.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 5.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.1 | 1.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 2.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.1 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.0 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.4 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 5.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 4.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.6 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.7 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.6 | GO:0009986 | cell surface(GO:0009986) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.6 | 6.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.4 | 4.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 1.1 | 7.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.0 | 4.1 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.9 | 2.8 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.9 | 2.8 | GO:0004753 | saccharopine dehydrogenase activity(GO:0004753) saccharopine dehydrogenase (NAD+, L-glutamate-forming) activity(GO:0047131) |
| 0.9 | 4.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.9 | 2.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.7 | 5.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.7 | 2.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.6 | 2.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.6 | 3.0 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.5 | 2.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.5 | 1.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.6 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.5 | 3.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.5 | 2.6 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.5 | 1.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 2.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 1.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.4 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 1.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.4 | 1.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.4 | 1.6 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.4 | 6.0 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.4 | 1.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.3 | 5.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 3.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.3 | 1.3 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.2 | GO:0003823 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.3 | 1.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 2.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.2 | 4.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.2 | 2.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 0.6 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.2 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 4.9 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.2 | 2.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 9.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 1.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 0.4 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 1.6 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 4.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 3.3 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 1.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.8 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 2.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 4.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.3 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.1 | 8.0 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 2.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 2.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 7.4 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.1 | 1.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 2.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 4.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 6.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.8 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.5 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 2.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 3.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 1.9 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 0.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 4.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 5.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.3 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.2 | 10.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 4.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 5.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.7 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 3.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 3.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 2.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.4 | 4.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 7.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.4 | 3.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.3 | 4.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 2.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.2 | 3.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 3.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 5.2 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 1.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 3.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.3 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 2.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 3.0 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 4.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |