Project
DANIO-CODE
Navigation
Downloads
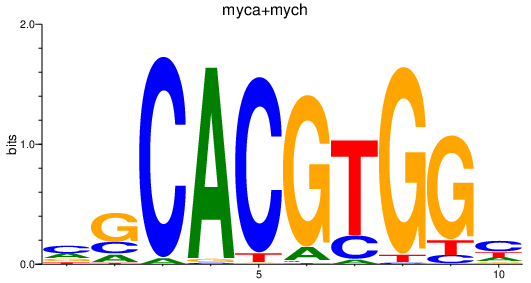
Results for myca+mych
Z-value: 1.28
Transcription factors associated with myca+mych
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myca
|
ENSDARG00000045695 | MYC proto-oncogene, bHLH transcription factor a |
|
mych
|
ENSDARG00000077473 | myelocytomatosis oncogene homolog |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myca | dr10_dc_chr24_+_10273081_10273133 | -0.77 | 5.1e-04 | Click! |
| mych | dr10_dc_chr6_+_50452229_50452254 | 0.50 | 4.6e-02 | Click! |
Activity profile of myca+mych motif
Sorted Z-values of myca+mych motif
Network of associatons between targets according to the STRING database.
First level regulatory network of myca+mych
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_14308032 | 4.40 |
|
|
|
| chr9_-_12916555 | 3.89 |
ENSDART00000140691
|
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr25_+_17593164 | 3.83 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr25_+_17593080 | 3.56 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr12_+_13053552 | 3.26 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr10_+_22065599 | 2.94 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr5_-_15971338 | 2.92 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr21_-_2296253 | 2.90 |
ENSDART00000162867
|
zgc:66483
|
zgc:66483 |
| chr22_-_28828375 | 2.88 |
ENSDART00000104880
ENSDART00000005112 |
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr15_-_17163526 | 2.64 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr5_-_31738565 | 2.49 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr23_-_21607742 | 2.33 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr22_+_15317622 | 2.29 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr17_+_15527923 | 2.21 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr16_-_31834830 | 2.21 |
|
|
|
| chr25_+_17593235 | 2.19 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr14_+_15845853 | 2.06 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr7_+_30355221 | 2.05 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr4_-_14193396 | 2.03 |
ENSDART00000101812
ENSDART00000143804 |
pus7l
|
pseudouridylate synthase 7-like |
| chr18_+_14677042 | 2.02 |
ENSDART00000138995
|
vps9d1
|
VPS9 domain containing 1 |
| chr22_+_17235696 | 1.91 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr19_-_24971920 | 1.89 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr13_-_15011369 | 1.89 |
ENSDART00000163132
|
rab11fip5a
|
RAB11 family interacting protein 5a (class I) |
| chr22_+_2237813 | 1.86 |
|
|
|
| chr19_+_39689450 | 1.78 |
|
|
|
| chr23_+_38335290 | 1.72 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr13_-_44885278 | 1.71 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr8_+_25276000 | 1.67 |
ENSDART00000049793
|
gstm.1
|
glutathione S-transferase mu, tandem duplicate 1 |
| KN150266v1_-_68652 | 1.63 |
|
|
|
| chr24_+_34183462 | 1.62 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr21_-_30045531 | 1.62 |
ENSDART00000157307
|
ccnjl
|
cyclin J-like |
| chr25_+_21001126 | 1.58 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr19_+_21209328 | 1.57 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr5_-_19619201 | 1.52 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr13_+_31271568 | 1.51 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr22_+_4734033 | 1.51 |
ENSDART00000158846
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr16_-_31834774 | 1.51 |
|
|
|
| chr10_-_14971401 | 1.50 |
ENSDART00000044756
ENSDART00000128579 |
smad2
|
SMAD family member 2 |
| chr4_-_13025193 | 1.50 |
ENSDART00000177894
|
BX119963.3
|
ENSDARG00000106390 |
| chr11_-_44859225 | 1.49 |
ENSDART00000163776
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr8_+_40126740 | 1.47 |
|
|
|
| chr24_+_39631194 | 1.45 |
ENSDART00000013894
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr5_-_19619115 | 1.44 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr21_-_31215463 | 1.43 |
ENSDART00000170507
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr10_-_14971565 | 1.42 |
ENSDART00000147653
|
smad2
|
SMAD family member 2 |
| chr22_+_4733940 | 1.42 |
ENSDART00000158279
|
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr18_-_11626694 | 1.41 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr20_+_38354928 | 1.41 |
ENSDART00000149160
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr23_+_38228263 | 1.39 |
ENSDART00000137969
|
zgc:112994
|
zgc:112994 |
| chr21_+_25031903 | 1.37 |
|
|
|
| chr1_-_55072271 | 1.35 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr20_-_47994260 | 1.33 |
ENSDART00000166857
|
CABZ01059120.1
|
ENSDARG00000100162 |
| chr23_+_30803887 | 1.33 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr13_-_29571309 | 1.32 |
ENSDART00000139773
|
CT025909.1
|
ENSDARG00000093614 |
| chr25_+_33118422 | 1.32 |
|
|
|
| chr11_+_25058986 | 1.31 |
ENSDART00000122249
|
top1l
|
topoisomerase (DNA) I, like |
| chr13_-_2296283 | 1.29 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr5_+_68036497 | 1.26 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr1_+_47323244 | 1.26 |
|
|
|
| chr8_-_44247277 | 1.25 |
|
|
|
| chr23_+_30803518 | 1.24 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr3_-_18655432 | 1.22 |
ENSDART00000034489
|
msrb1a
|
methionine sulfoxide reductase B1a |
| chr11_+_6012910 | 1.21 |
ENSDART00000161001
|
gtpbp3
|
GTP binding protein 3 |
| chr8_-_38284748 | 1.21 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr13_+_35799681 | 1.21 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr13_+_30441704 | 1.21 |
ENSDART00000010052
|
ppifa
|
peptidylprolyl isomerase Fa |
| chr1_+_40134345 | 1.19 |
ENSDART00000177517
|
CR387992.1
|
ENSDARG00000108790 |
| chr10_+_43953171 | 1.18 |
|
|
|
| chr24_+_34183557 | 1.18 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr5_-_27394361 | 1.17 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr3_+_18657831 | 1.16 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr10_-_39362315 | 1.15 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr3_+_41984648 | 1.14 |
ENSDART00000154052
|
ftsj2
|
FtsJ RNA methyltransferase homolog 2 (E. coli) |
| chr14_+_15845943 | 1.14 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr1_-_18955249 | 1.13 |
ENSDART00000146688
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr7_-_31346802 | 1.13 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr11_-_13094557 | 1.13 |
ENSDART00000160989
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr13_-_11888944 | 1.11 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr21_+_6009129 | 1.10 |
ENSDART00000065858
|
fpgs
|
folylpolyglutamate synthase |
| chr1_-_29958364 | 1.10 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr15_-_1519615 | 1.10 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr19_-_34529880 | 1.10 |
ENSDART00000158677
ENSDART00000112004 |
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr6_+_3556296 | 1.09 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr5_-_9577533 | 1.09 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr13_-_25277861 | 1.09 |
ENSDART00000002741
|
itprip
|
inositol 1,4,5-trisphosphate receptor interacting protein |
| chr24_+_39630741 | 1.09 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr24_+_36429525 | 1.07 |
ENSDART00000062722
|
pus3
|
pseudouridylate synthase 3 |
| chr23_+_21608152 | 1.07 |
|
|
|
| KN149704v1_-_17031 | 1.06 |
|
|
|
| chr21_-_22085596 | 1.06 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr1_-_39297624 | 1.05 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr19_-_9020995 | 1.05 |
ENSDART00000043507
|
ciarta
|
circadian associated repressor of transcription a |
| chr2_+_44928188 | 1.05 |
ENSDART00000147292
|
ece2a
|
endothelin converting enzyme 2a |
| chr15_-_28852613 | 1.04 |
ENSDART00000152147
|
blmh
|
bleomycin hydrolase |
| chr9_-_27585644 | 1.04 |
ENSDART00000101401
|
tex30
|
testis expressed 30 |
| chr9_-_904565 | 1.04 |
ENSDART00000144068
|
zgc:101851
|
zgc:101851 |
| chr20_-_46079578 | 1.03 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr13_-_44885345 | 1.03 |
ENSDART00000074787
ENSDART00000125633 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr11_+_25058714 | 1.03 |
ENSDART00000065949
|
top1l
|
topoisomerase (DNA) I, like |
| chr17_+_22740348 | 1.02 |
ENSDART00000151999
|
ttc27
|
tetratricopeptide repeat domain 27 |
| chr22_-_26971170 | 1.02 |
ENSDART00000162742
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr11_+_24562450 | 1.02 |
ENSDART00000082761
ENSDART00000131976 |
adipor1a
|
adiponectin receptor 1a |
| chr3_-_5157662 | 1.02 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr21_+_21760105 | 1.02 |
ENSDART00000003518
|
neu3.2
|
sialidase 3 (membrane sialidase), tandem duplicate 2 |
| chr18_+_8959686 | 1.00 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr5_-_12592887 | 1.00 |
|
|
|
| chr20_-_25732204 | 1.00 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr8_+_10825036 | 1.00 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr25_+_18997440 | 1.00 |
ENSDART00000155569
|
si:dkeyp-19h3.8
|
si:dkeyp-19h3.8 |
| chr8_-_38284959 | 0.99 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr23_-_33396673 | 0.98 |
|
|
|
| chr22_-_23252993 | 0.97 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr21_+_25032002 | 0.96 |
|
|
|
| chr15_-_43954665 | 0.96 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr16_+_17857553 | 0.96 |
ENSDART00000148878
|
them4
|
thioesterase superfamily member 4 |
| chr3_-_40791555 | 0.96 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr9_-_27581087 | 0.95 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr3_+_50398735 | 0.95 |
|
|
|
| chr8_-_38284904 | 0.94 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr20_+_25726978 | 0.94 |
ENSDART00000164264
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr7_-_31346844 | 0.92 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr9_+_42656327 | 0.92 |
|
|
|
| chr12_-_33256599 | 0.92 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_33874289 | 0.92 |
ENSDART00000125745
|
sfxn2
|
sideroflexin 2 |
| chr13_+_30441803 | 0.91 |
ENSDART00000144417
|
ppifa
|
peptidylprolyl isomerase Fa |
| chr16_+_41004372 | 0.91 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr3_-_40790889 | 0.91 |
|
|
|
| chr14_-_5100304 | 0.91 |
ENSDART00000168074
|
pcgf1
|
polycomb group ring finger 1 |
| chr18_+_6498590 | 0.91 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr23_+_38335341 | 0.91 |
ENSDART00000177981
ENSDART00000178842 |
CABZ01070579.1
|
ENSDARG00000109193 |
| chr3_+_41984196 | 0.91 |
ENSDART00000154052
|
ftsj2
|
FtsJ RNA methyltransferase homolog 2 (E. coli) |
| chr19_-_22761674 | 0.90 |
|
|
|
| chr5_+_60782855 | 0.90 |
ENSDART00000074073
|
alkbh4
|
alkB homolog 4, lysine demthylase |
| chr17_+_26785230 | 0.90 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr4_-_3015097 | 0.90 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr20_-_33802082 | 0.88 |
ENSDART00000166573
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr3_-_40791490 | 0.87 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr23_+_38313746 | 0.87 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr6_-_44164161 | 0.86 |
ENSDART00000035513
|
shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr10_+_29883772 | 0.86 |
ENSDART00000100032
|
hyou1
|
hypoxia up-regulated 1 |
| chr13_-_35781185 | 0.86 |
|
|
|
| chr21_+_34085267 | 0.85 |
ENSDART00000024750
|
hmgb3b
|
high mobility group box 3b |
| chr24_+_32754013 | 0.85 |
ENSDART00000156638
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| chr5_-_27394324 | 0.85 |
ENSDART00000132740
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr14_-_21779536 | 0.84 |
|
|
|
| chr23_-_21608024 | 0.84 |
ENSDART00000134587
|
rcc2
|
regulator of chromosome condensation 2 |
| chr1_-_39297686 | 0.83 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr14_-_35932521 | 0.83 |
ENSDART00000158722
|
BX511223.1
|
ENSDARG00000101064 |
| chr21_+_25031760 | 0.82 |
|
|
|
| chr3_+_43361682 | 0.81 |
ENSDART00000168784
|
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr1_+_47323043 | 0.81 |
|
|
|
| chr23_-_21608362 | 0.81 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr25_+_17593263 | 0.80 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr21_-_11235148 | 0.80 |
ENSDART00000122331
|
rtkn2b
|
rhotekin 2b |
| chr8_+_42910943 | 0.79 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr23_+_38314255 | 0.78 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr12_-_28422582 | 0.78 |
ENSDART00000067762
|
MYO1D
|
myosin ID |
| chr17_-_31802796 | 0.76 |
ENSDART00000172519
|
abraxas2b
|
abraxas 2b, BRISC complex subunit |
| chr24_-_38304083 | 0.76 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr20_-_7079663 | 0.76 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr21_-_30045669 | 0.76 |
ENSDART00000155188
|
ccnjl
|
cyclin J-like |
| chr21_+_21760059 | 0.75 |
ENSDART00000003518
|
neu3.2
|
sialidase 3 (membrane sialidase), tandem duplicate 2 |
| chr15_+_17163165 | 0.75 |
|
|
|
| chr22_-_28828300 | 0.75 |
ENSDART00000104880
|
si:dkeyp-34c12.1
|
si:dkeyp-34c12.1 |
| chr13_-_11888896 | 0.75 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr22_+_15317535 | 0.75 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr13_+_39151166 | 0.75 |
ENSDART00000113259
|
ENSDARG00000079055
|
ENSDARG00000079055 |
| chr20_-_7079412 | 0.74 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr4_+_14107347 | 0.74 |
ENSDART00000150748
|
CU469420.1
|
ENSDARG00000086259 |
| chr14_+_41042379 | 0.74 |
ENSDART00000173335
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr3_+_41984701 | 0.73 |
ENSDART00000154052
|
ftsj2
|
FtsJ RNA methyltransferase homolog 2 (E. coli) |
| chr3_+_5125139 | 0.73 |
ENSDART00000161755
|
CU681842.1
|
ENSDARG00000102795 |
| chr17_+_15528557 | 0.73 |
|
|
|
| chr21_-_31215400 | 0.72 |
ENSDART00000121946
|
crcp
|
calcitonin gene-related peptide-receptor component protein |
| chr5_-_67315284 | 0.72 |
ENSDART00000131665
|
gtf3aa
|
general transcription factor IIIAa |
| chr7_-_55238300 | 0.72 |
ENSDART00000148514
|
aprt
|
adenine phosphoribosyltransferase |
| chr8_+_26413471 | 0.71 |
ENSDART00000172542
ENSDART00000078369 |
zgc:136971
|
zgc:136971 |
| chr15_+_17162691 | 0.71 |
|
|
|
| chr3_-_40791035 | 0.70 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr9_+_27909662 | 0.70 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr13_-_35781541 | 0.70 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr8_+_26040518 | 0.69 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr23_-_38228511 | 0.69 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr13_-_35766296 | 0.69 |
ENSDART00000162399
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr16_-_13723778 | 0.69 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr18_-_20618924 | 0.68 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr12_-_5153412 | 0.68 |
ENSDART00000160037
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr11_-_44859336 | 0.68 |
ENSDART00000157615
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr21_+_21760026 | 0.68 |
ENSDART00000003518
|
neu3.2
|
sialidase 3 (membrane sialidase), tandem duplicate 2 |
| chr22_+_17235538 | 0.67 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| KN150266v1_-_68620 | 0.67 |
|
|
|
| chr18_+_5637539 | 0.67 |
|
|
|
| chr13_-_35781256 | 0.67 |
|
|
|
| chr9_-_21649644 | 0.67 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr24_+_35297193 | 0.65 |
ENSDART00000075142
ENSDART00000158801 |
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr24_+_35296732 | 0.65 |
ENSDART00000172652
|
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr19_-_22762165 | 0.64 |
ENSDART00000169065
ENSDART00000080260 |
zgc:109744
|
zgc:109744 |
| chr3_+_43361636 | 0.62 |
ENSDART00000161277
|
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr9_-_27909272 | 0.62 |
ENSDART00000161068
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr17_+_26785073 | 0.61 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.8 | 2.5 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.7 | 2.2 | GO:2001284 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.7 | 2.7 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.7 | 3.3 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.6 | 2.5 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.5 | 4.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.5 | 1.4 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.4 | 1.3 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.4 | 2.6 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.4 | 4.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.4 | 1.7 | GO:0051977 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.4 | 2.9 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.4 | 2.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.4 | 1.5 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.4 | 1.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.4 | 2.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 1.9 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 0.9 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.3 | 1.4 | GO:0002548 | monocyte chemotaxis(GO:0002548) response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.3 | 1.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.3 | 1.1 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 1.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.2 | 2.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.5 | GO:0034475 | U4 snRNA 3'-end processing(GO:0034475) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.2 | 2.5 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.2 | 2.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.4 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 2.0 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 0.4 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.6 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.1 | 0.6 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.3 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 1.1 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.5 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 2.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.8 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.6 | GO:0021571 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.1 | 3.1 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 0.8 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.9 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.1 | 1.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 4.4 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 2.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 2.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.6 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.1 | 1.7 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 1.9 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.9 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 3.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.1 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.8 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 10.4 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.2 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 1.0 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.1 | GO:0045226 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.0 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 1.4 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.0 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.8 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 1.6 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.1 | GO:0046112 | nucleobase biosynthetic process(GO:0046112) |
| 0.0 | 0.2 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 1.9 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 1.3 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.6 | 2.9 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 2.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.5 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 1.9 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.3 | 2.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.3 | 4.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 4.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.6 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.7 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 2.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 3.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 2.0 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.5 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 5.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 4.0 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 1.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 4.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.9 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 3.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 10.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 4.6 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 3.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.6 | GO:0000922 | spindle pole(GO:0000922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.6 | 2.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.6 | 1.9 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.6 | 2.5 | GO:0061697 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.6 | 2.9 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.4 | 1.7 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.4 | 1.2 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.4 | 2.4 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.4 | 1.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.4 | 1.5 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 0.4 | 1.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 1.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 0.7 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.2 | 1.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 2.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 0.8 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 2.1 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 6.8 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.2 | 1.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 3.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 2.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 2.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 3.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.3 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.8 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.4 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 2.1 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.8 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 1.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 4.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 3.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 0.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 4.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 2.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 3.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 3.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 3.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 2.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 1.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 2.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 2.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 3.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.1 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 4.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |