Project
DANIO-CODE
Navigation
Downloads
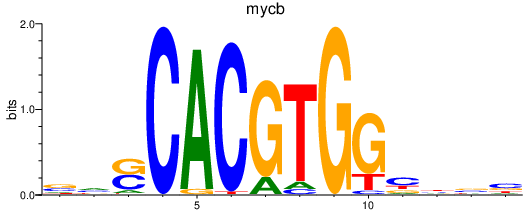
Results for mycb
Z-value: 1.42
Transcription factors associated with mycb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mycb
|
ENSDARG00000007241 | MYC proto-oncogene, bHLH transcription factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mycb | dr10_dc_chr2_+_32033176_32033202 | -0.54 | 3.1e-02 | Click! |
Activity profile of mycb motif
Sorted Z-values of mycb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of mycb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_24971633 | 6.72 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr20_-_23184142 | 4.64 |
ENSDART00000176282
|
CU693368.2
|
ENSDARG00000108718 |
| chr15_+_19902697 | 3.04 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr16_+_24045774 | 3.03 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr8_+_46378250 | 2.96 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr24_+_23599223 | 2.53 |
|
|
|
| chr11_+_6446302 | 2.47 |
ENSDART00000140707
ENSDART00000036939 |
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr23_-_28099284 | 2.22 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr11_+_7139675 | 2.11 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr3_-_23444371 | 2.03 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr19_-_24971920 | 1.92 |
ENSDART00000132660
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr1_+_16683931 | 1.89 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr11_-_18393802 | 1.85 |
ENSDART00000125453
|
dido1
|
death inducer-obliterator 1 |
| chr10_+_4962403 | 1.84 |
ENSDART00000134679
|
CU074419.2
|
ENSDARG00000093688 |
| chr5_-_8260713 | 1.78 |
ENSDART00000167793
|
mybbp1a
|
MYB binding protein (P160) 1a |
| chr7_+_24770873 | 1.76 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr22_-_570067 | 1.76 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr20_+_54289892 | 1.70 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr6_-_39656043 | 1.69 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr12_-_7572970 | 1.65 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr14_+_241493 | 1.57 |
ENSDART00000002232
|
msx1a
|
muscle segment homeobox 1a |
| chr11_-_18027225 | 1.56 |
|
|
|
| chr22_-_26971466 | 1.50 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr16_+_24045707 | 1.47 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr4_-_876235 | 1.44 |
|
|
|
| chr17_+_53068731 | 1.43 |
ENSDART00000156774
|
dph6
|
diphthamine biosynthesis 6 |
| chr20_-_25732091 | 1.43 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr23_-_28099359 | 1.42 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr19_-_24971335 | 1.37 |
ENSDART00000176022
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr17_-_48623315 | 1.35 |
ENSDART00000030934
|
kcnk5a
|
potassium channel, subfamily K, member 5a |
| chr11_+_3940085 | 1.35 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr19_-_35642440 | 1.34 |
ENSDART00000167853
ENSDART00000054274 |
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr11_-_40415291 | 1.34 |
|
|
|
| chr23_-_28099235 | 1.33 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr3_-_12032297 | 1.32 |
ENSDART00000140123
|
hmox2b
|
heme oxygenase 2b |
| chr3_+_6150498 | 1.30 |
ENSDART00000175915
|
BX284638.2
|
ENSDARG00000108888 |
| chr19_+_7233537 | 1.27 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| KN150670v1_-_32201 | 1.24 |
|
|
|
| chr20_-_25726868 | 1.19 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr6_-_39655998 | 1.19 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr23_-_28099391 | 1.19 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr8_-_46378116 | 1.17 |
ENSDART00000136602
|
qars
|
glutaminyl-tRNA synthetase |
| chr8_+_46378706 | 1.15 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr10_-_22065636 | 1.15 |
|
|
|
| chr17_+_19479310 | 1.13 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr23_-_38228305 | 1.13 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr5_+_68036497 | 1.11 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr3_-_55995924 | 1.11 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr9_+_907908 | 1.06 |
ENSDART00000144114
|
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr6_-_39083081 | 1.06 |
ENSDART00000021520
|
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr9_+_38348762 | 1.04 |
ENSDART00000059549
|
nifk
|
nucleolar protein interacting with the FHA domain of MKI67 |
| chr16_+_5466342 | 1.02 |
ENSDART00000160008
|
plecb
|
plectin b |
| chr23_-_21607811 | 0.98 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr2_+_19339251 | 0.96 |
ENSDART00000172148
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr20_+_25546646 | 0.95 |
ENSDART00000012581
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr19_-_27379910 | 0.94 |
ENSDART00000103955
|
zbtb12.2
|
zinc finger and BTB domain containing 12, tandem duplicate 2 |
| chr19_+_40661722 | 0.92 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr23_+_23559246 | 0.92 |
ENSDART00000172214
|
agrn
|
agrin |
| chr12_-_34614931 | 0.90 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr19_-_25173832 | 0.90 |
ENSDART00000161016
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr11_-_2107054 | 0.90 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr16_-_38679062 | 0.90 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr23_+_36010592 | 0.89 |
ENSDART00000137507
|
hoxc3a
|
homeo box C3a |
| chr10_+_29964045 | 0.89 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr11_+_3939876 | 0.88 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| KN150670v1_-_32341 | 0.87 |
|
|
|
| chr2_-_23431625 | 0.85 |
ENSDART00000157318
|
BX548024.1
|
ENSDARG00000097593 |
| chr13_-_25068717 | 0.85 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr21_+_26035166 | 0.85 |
ENSDART00000134939
|
rpl23a
|
ribosomal protein L23a |
| chr20_-_20412830 | 0.84 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr22_-_37676556 | 0.83 |
ENSDART00000028085
|
ttc14
|
tetratricopeptide repeat domain 14 |
| chr9_+_33979089 | 0.83 |
ENSDART00000144623
|
kdm6a
|
lysine (K)-specific demethylase 6A |
| chr13_-_35781888 | 0.82 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr2_-_45375072 | 0.81 |
|
|
|
| chr9_-_21649243 | 0.80 |
ENSDART00000133469
|
zmym2
|
zinc finger, MYM-type 2 |
| chr19_-_27379989 | 0.80 |
ENSDART00000103955
|
zbtb12.2
|
zinc finger and BTB domain containing 12, tandem duplicate 2 |
| chr10_+_38452427 | 0.80 |
|
|
|
| chr4_-_15208571 | 0.80 |
|
|
|
| chr21_+_38591263 | 0.80 |
ENSDART00000143373
|
rbmx2
|
RNA binding motif protein, X-linked 2 |
| chr13_-_35782030 | 0.77 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr25_+_17308788 | 0.77 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr25_-_8436134 | 0.77 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr4_+_377829 | 0.76 |
ENSDART00000030215
|
rpl18a
|
ribosomal protein L18a |
| chr8_-_2187555 | 0.75 |
ENSDART00000135835
|
si:dkeyp-117b11.3
|
si:dkeyp-117b11.3 |
| chr10_+_22066137 | 0.73 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr24_-_23155932 | 0.71 |
|
|
|
| chr19_+_40661998 | 0.70 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr13_-_9035530 | 0.70 |
ENSDART00000138961
|
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr9_-_11705194 | 0.69 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr15_-_14616083 | 0.68 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_+_13088982 | 0.68 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr19_-_39048402 | 0.67 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr9_+_50670635 | 0.67 |
ENSDART00000179475
|
CABZ01083707.1
|
ENSDARG00000107278 |
| chr4_-_3015097 | 0.66 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr24_-_42120772 | 0.65 |
ENSDART00000166413
ENSDART00000166414 |
ssr1
|
signal sequence receptor, alpha |
| chr12_+_42281025 | 0.65 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr21_-_45341242 | 0.65 |
ENSDART00000075438
|
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr16_+_17061069 | 0.64 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr10_+_22066361 | 0.64 |
|
|
|
| chr4_+_15208675 | 0.62 |
ENSDART00000148049
|
BX537358.1
|
ENSDARG00000093247 |
| chr21_-_11877044 | 0.59 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr11_-_42826308 | 0.58 |
|
|
|
| chr13_-_11887665 | 0.58 |
|
|
|
| chr15_-_44029439 | 0.57 |
|
|
|
| chr21_-_30371923 | 0.56 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr8_-_2187590 | 0.56 |
ENSDART00000135835
|
si:dkeyp-117b11.3
|
si:dkeyp-117b11.3 |
| chr1_-_54294303 | 0.56 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr21_+_17731439 | 0.55 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr16_+_24045675 | 0.55 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr15_+_6655082 | 0.55 |
ENSDART00000157678
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr13_-_35781185 | 0.55 |
|
|
|
| chr19_+_7838616 | 0.54 |
ENSDART00000178973
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr20_-_47805830 | 0.53 |
ENSDART00000097888
|
CABZ01059104.1
|
ENSDARG00000067927 |
| chr18_-_5235925 | 0.53 |
ENSDART00000082087
|
nip7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr14_+_51692872 | 0.53 |
ENSDART00000163856
|
noa1
|
nitric oxide associated 1 |
| chr7_+_58718614 | 0.53 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr2_+_19587617 | 0.53 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr9_+_42019770 | 0.52 |
ENSDART00000097295
|
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr2_-_42525768 | 0.51 |
ENSDART00000026339
|
gtpbp4
|
GTP binding protein 4 |
| chr23_+_25209497 | 0.51 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
| chr20_+_19175518 | 0.51 |
|
|
|
| chr13_-_44885345 | 0.50 |
ENSDART00000074787
ENSDART00000125633 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr14_+_28126921 | 0.50 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr9_-_12681592 | 0.50 |
ENSDART00000052256
|
sumo3b
|
small ubiquitin-like modifier 3b |
| chr19_-_791149 | 0.50 |
ENSDART00000151782
ENSDART00000037515 |
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr1_-_5796394 | 0.49 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr2_+_39653315 | 0.49 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr10_+_24476213 | 0.48 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_-_39048324 | 0.48 |
ENSDART00000086717
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr12_-_9662735 | 0.48 |
ENSDART00000161063
|
heatr1
|
HEAT repeat containing 1 |
| chr20_-_30474196 | 0.47 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr11_+_25248195 | 0.46 |
ENSDART00000026249
|
gnl3l
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr19_+_42758938 | 0.45 |
ENSDART00000077059
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr13_+_30781652 | 0.44 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr25_+_29999307 | 0.44 |
ENSDART00000003346
|
pdcd2l
|
programmed cell death 2-like |
| chr11_+_25058714 | 0.42 |
ENSDART00000065949
|
top1l
|
topoisomerase (DNA) I, like |
| chr24_+_9603813 | 0.42 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr19_-_2222108 | 0.41 |
ENSDART00000127578
|
twistnb
|
TWIST neighbor |
| chr2_-_10602948 | 0.40 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr4_+_18975027 | 0.39 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr13_-_35781541 | 0.37 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr24_+_16760759 | 0.36 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr13_-_2296283 | 0.36 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr19_+_20617339 | 0.35 |
ENSDART00000133633
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr1_-_54294382 | 0.35 |
ENSDART00000038330
|
khsrp
|
KH-type splicing regulatory protein |
| chr3_-_39245184 | 0.35 |
|
|
|
| chr14_+_15982295 | 0.34 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr10_+_29964164 | 0.34 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr20_+_54463948 | 0.34 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr1_+_44099012 | 0.33 |
ENSDART00000073712
|
tmem187
|
transmembrane protein 187 |
| chr23_-_21608362 | 0.33 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr1_-_54294205 | 0.33 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr23_+_25209273 | 0.33 |
ENSDART00000103986
|
fam3a
|
family with sequence similarity 3, member A |
| chr1_-_43323814 | 0.33 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr2_+_16492315 | 0.33 |
ENSDART00000135783
|
selj
|
selenoprotein J |
| chr24_+_32754013 | 0.32 |
ENSDART00000156638
|
si:ch211-282b22.1
|
si:ch211-282b22.1 |
| KN149932v1_+_27584 | 0.31 |
|
|
|
| chr3_+_25247071 | 0.31 |
|
|
|
| chr6_+_59776364 | 0.31 |
ENSDART00000048449
ENSDART00000160405 |
mtrf1
|
mitochondrial translational release factor 1 |
| chr9_+_30174202 | 0.31 |
ENSDART00000020743
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr10_+_39362458 | 0.31 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr21_-_11877525 | 0.30 |
ENSDART00000145194
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr9_+_15281351 | 0.30 |
|
|
|
| chr9_-_46609148 | 0.28 |
|
|
|
| chr13_-_44885278 | 0.28 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr10_+_29963998 | 0.27 |
ENSDART00000118838
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr13_+_30781718 | 0.25 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr3_-_55995563 | 0.25 |
|
|
|
| chr5_-_31275147 | 0.24 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr14_+_5078937 | 0.24 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr5_+_3604487 | 0.23 |
ENSDART00000030125
|
znhit3
|
zinc finger, HIT-type containing 3 |
| chr16_-_21863723 | 0.23 |
ENSDART00000161640
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chr2_+_22196842 | 0.22 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr24_-_39630728 | 0.22 |
ENSDART00000031486
|
lyrm1
|
LYR motif containing 1 |
| chr23_-_21607742 | 0.21 |
ENSDART00000139092
|
rcc2
|
regulator of chromosome condensation 2 |
| chr21_-_30371822 | 0.21 |
ENSDART00000138384
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr20_+_54464026 | 0.21 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr24_-_16760535 | 0.21 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr25_+_5567388 | 0.20 |
|
|
|
| chr20_+_4078243 | 0.20 |
ENSDART00000124197
|
CU929133.1
|
ENSDARG00000088444 |
| chr21_-_11106675 | 0.19 |
ENSDART00000167666
|
dnajc21
|
DnaJ (Hsp40) homolog, subfamily C, member 21 |
| chr11_-_42826276 | 0.19 |
|
|
|
| chr3_-_45836485 | 0.18 |
|
|
|
| chr20_+_34768464 | 0.18 |
ENSDART00000152836
|
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr13_+_31271568 | 0.18 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr12_+_35849423 | 0.18 |
|
|
|
| chr23_-_17174825 | 0.17 |
ENSDART00000053418
|
dnmt3bb.1
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.1 |
| chr21_+_4345107 | 0.17 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr22_+_17235538 | 0.17 |
ENSDART00000134798
|
tdrd5
|
tudor domain containing 5 |
| chr1_-_25838798 | 0.17 |
|
|
|
| chr5_-_54091732 | 0.16 |
ENSDART00000150070
|
ccnb1
|
cyclin B1 |
| chr13_-_2296041 | 0.16 |
ENSDART00000126697
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr13_-_35781256 | 0.16 |
|
|
|
| chr7_+_52612436 | 0.16 |
ENSDART00000109973
|
tp53bp1
|
tumor protein p53 binding protein, 1 |
| chr14_+_21779325 | 0.16 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr22_+_10683985 | 0.15 |
ENSDART00000063207
ENSDART00000122349 |
hiat1b
|
hippocampus abundant transcript 1b |
| chr21_+_3645203 | 0.15 |
ENSDART00000099535
|
spout1
|
SPOUT domain containing methyltransferase 1 |
| chr2_+_10148394 | 0.15 |
ENSDART00000158214
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr4_-_1989775 | 0.15 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr21_-_4086050 | 0.15 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.8 | 2.5 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 0.4 | 1.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.4 | 3.0 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.4 | 4.1 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.4 | 1.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.3 | 1.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.3 | 9.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.3 | 1.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 1.6 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.2 | 2.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.9 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.2 | 0.8 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.2 | 0.8 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.7 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 | 0.3 | GO:1901797 | negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.2 | 2.2 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.2 | 6.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.4 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.1 | 1.8 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.3 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.1 | 0.4 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) Wnt protein secretion(GO:0061355) |
| 0.1 | 0.8 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.1 | 0.5 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.8 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.1 | 1.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.6 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.4 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.7 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 1.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.5 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 2.9 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.8 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 1.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 9.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.3 | 1.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.3 | 0.9 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.3 | 2.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 0.8 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.2 | 0.8 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.8 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.5 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 7.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.2 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.0 | 4.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.5 | 2.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 1.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.3 | 1.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 0.9 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.2 | 0.8 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.2 | 0.5 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 0.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 0.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.0 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.8 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.8 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 2.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 3.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 4.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.8 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 4.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 1.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 1.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.2 | 1.8 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 3.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.8 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |