Project
DANIO-CODE
Navigation
Downloads
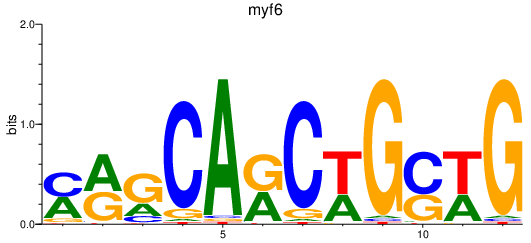
Results for myf6
Z-value: 1.74
Transcription factors associated with myf6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myf6
|
ENSDARG00000029830 | myogenic factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myf6 | dr10_dc_chr4_+_21996784_21996805 | -0.72 | 1.5e-03 | Click! |
Activity profile of myf6 motif
Sorted Z-values of myf6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of myf6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_73890950 | 6.41 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr7_-_73890817 | 4.56 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr7_-_59284273 | 3.97 |
ENSDART00000008554
|
larp7
|
La ribonucleoprotein domain family, member 7 |
| chr7_-_59284362 | 3.75 |
ENSDART00000101731
|
larp7
|
La ribonucleoprotein domain family, member 7 |
| chr19_-_27093095 | 3.23 |
|
|
|
| chr1_-_30299017 | 2.90 |
|
|
|
| chr1_-_44892852 | 2.43 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr12_+_30673985 | 2.38 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr6_+_135002 | 2.31 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr7_+_17848688 | 2.28 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr9_+_22846286 | 2.23 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr4_-_6443552 | 2.18 |
ENSDART00000136572
|
foxp2
|
forkhead box P2 |
| chr2_+_28199458 | 2.18 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr1_-_44892600 | 2.15 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr23_+_46000362 | 2.14 |
ENSDART00000023944
|
lmnl3
|
lamin L3 |
| chr23_-_6831711 | 2.12 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr14_-_7102535 | 2.08 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr20_-_9212139 | 2.07 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr21_-_38670059 | 2.05 |
ENSDART00000065169
ENSDART00000113813 |
siah2l
|
seven in absentia homolog 2 (Drosophila)-like |
| chr2_-_39761055 | 1.99 |
ENSDART00000179646
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr12_-_10474942 | 1.92 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr9_-_32240567 | 1.90 |
|
|
|
| chr17_+_699708 | 1.89 |
ENSDART00000165144
|
siva1
|
SIVA1, apoptosis-inducing factor |
| KN150455v1_-_14200 | 1.86 |
|
|
|
| chr20_-_14885629 | 1.85 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr1_-_39622531 | 1.84 |
|
|
|
| chr12_-_24711074 | 1.74 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr2_-_54224744 | 1.72 |
|
|
|
| chr4_-_6443699 | 1.69 |
|
|
|
| chr19_+_14592632 | 1.69 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr13_+_11418060 | 1.67 |
ENSDART00000166908
|
desi2
|
desumoylating isopeptidase 2 |
| chr13_-_50299643 | 1.61 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr23_+_43868027 | 1.59 |
ENSDART00000112598
ENSDART00000169576 |
otud4
|
OTU deubiquitinase 4 |
| KN150589v1_-_5209 | 1.58 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr1_+_21964075 | 1.58 |
ENSDART00000161874
|
anxa5b
|
annexin A5b |
| chr25_-_34648876 | 1.53 |
ENSDART00000154851
|
zgc:153405
|
zgc:153405 |
| chr20_-_14885599 | 1.53 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr7_+_45747622 | 1.50 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr16_+_53391776 | 1.49 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr7_-_41446221 | 1.48 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr2_-_45810787 | 1.48 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr1_-_53052062 | 1.47 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr17_-_51729707 | 1.46 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr17_-_51729568 | 1.45 |
ENSDART00000157244
|
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr11_+_30005768 | 1.43 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr16_+_53391438 | 1.43 |
ENSDART00000010792
|
ptdss1a
|
phosphatidylserine synthase 1a |
| chr17_+_19606608 | 1.43 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr5_+_37144525 | 1.42 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr8_-_20106132 | 1.42 |
ENSDART00000029939
|
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr17_+_15527923 | 1.42 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr25_-_352985 | 1.41 |
ENSDART00000165705
|
szl
|
sizzled |
| chr3_-_31747486 | 1.41 |
ENSDART00000076189
|
zgc:171779
|
zgc:171779 |
| chr7_-_73890600 | 1.41 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr9_+_38832959 | 1.40 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr5_+_37144384 | 1.39 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr9_+_22846082 | 1.38 |
ENSDART00000139270
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr7_-_59284632 | 1.37 |
ENSDART00000101731
|
larp7
|
La ribonucleoprotein domain family, member 7 |
| chr5_-_41260390 | 1.35 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr7_-_49528331 | 1.35 |
ENSDART00000052083
|
fjx1
|
four jointed box 1 |
| chr8_+_25126238 | 1.35 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr8_+_28362546 | 1.35 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr2_-_58805035 | 1.34 |
ENSDART00000159735
|
mau2
|
MAU2 sister chromatid cohesion factor |
| chr25_-_20932941 | 1.34 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr16_+_25202230 | 1.33 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr18_+_37279099 | 1.32 |
ENSDART00000135444
|
tbcb
|
tubulin folding cofactor B |
| chr9_+_22846432 | 1.32 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr24_-_19574666 | 1.32 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr1_-_51075777 | 1.32 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr1_-_55072271 | 1.31 |
ENSDART00000142244
|
dnajb1b
|
DnaJ (Hsp40) homolog, subfamily B, member 1b |
| chr8_+_40463218 | 1.31 |
|
|
|
| chr9_-_18808049 | 1.31 |
|
|
|
| chr6_-_39315705 | 1.29 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr21_+_22598805 | 1.28 |
ENSDART00000142495
|
BX510940.1
|
ENSDARG00000089088 |
| chr1_+_41762057 | 1.27 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr12_-_24711169 | 1.26 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr20_-_3301951 | 1.25 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr17_+_28653205 | 1.24 |
ENSDART00000129779
|
hectd1
|
HECT domain containing 1 |
| chr13_+_11417882 | 1.24 |
ENSDART00000034935
|
desi2
|
desumoylating isopeptidase 2 |
| chr7_+_51520610 | 1.23 |
ENSDART00000174201
|
slc38a7
|
solute carrier family 38, member 7 |
| chr10_+_17919692 | 1.22 |
|
|
|
| chr20_-_38884093 | 1.21 |
ENSDART00000153430
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr17_-_6443618 | 1.21 |
ENSDART00000129950
ENSDART00000154074 |
ANKRD66
|
si:ch211-189e2.2 |
| chr24_-_42053682 | 1.20 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr15_+_14656797 | 1.19 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr12_-_25058837 | 1.19 |
ENSDART00000135368
|
rhoq
|
ras homolog family member Q |
| chr2_+_35134340 | 1.16 |
ENSDART00000113609
|
rfwd2
|
ring finger and WD repeat domain 2 |
| chr2_+_6341404 | 1.15 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr17_+_6310117 | 1.15 |
ENSDART00000149527
|
COQ8A (1 of many)
|
coenzyme Q8A |
| chr25_-_20933145 | 1.15 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| KN150455v1_-_14097 | 1.13 |
|
|
|
| chr9_-_29218911 | 1.12 |
ENSDART00000151941
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr3_-_15529158 | 1.11 |
ENSDART00000080441
|
zgc:66443
|
zgc:66443 |
| chr9_+_46748457 | 1.08 |
|
|
|
| chr5_-_35997198 | 1.08 |
ENSDART00000031270
|
rhogc
|
ras homolog gene family, member Gc |
| chr2_+_52225622 | 1.07 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr7_-_20330881 | 1.06 |
ENSDART00000169750
|
si:dkey-19b23.11
|
si:dkey-19b23.11 |
| KN150266v1_-_68652 | 1.06 |
|
|
|
| chr16_-_40633986 | 1.06 |
ENSDART00000170815
|
BX324007.2
|
ENSDARG00000100586 |
| chr2_-_31849916 | 1.06 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr24_-_23793117 | 1.05 |
|
|
|
| chr24_+_5771253 | 1.04 |
ENSDART00000154930
|
si:ch211-157j23.2
|
si:ch211-157j23.2 |
| chr6_-_3763783 | 1.04 |
ENSDART00000170584
|
tlk1b
|
tousled-like kinase 1b |
| chr7_-_41446119 | 1.03 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr18_+_45669615 | 1.02 |
ENSDART00000150973
|
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr20_-_3301898 | 1.02 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr19_+_41895061 | 1.02 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr7_+_24223131 | 1.01 |
ENSDART00000087568
|
nelfa
|
negative elongation factor complex member A |
| chr13_-_50299821 | 1.01 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr6_-_49160475 | 1.01 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr9_-_18807868 | 1.00 |
|
|
|
| chr5_-_45219194 | 1.00 |
|
|
|
| chr23_-_22596648 | 1.00 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr24_-_20496410 | 1.00 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr20_-_26516874 | 0.99 |
|
|
|
| chr24_-_19573966 | 0.99 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr7_+_34523283 | 0.98 |
ENSDART00000009698
|
esrp2
|
epithelial splicing regulatory protein 2 |
| chr20_+_35182783 | 0.98 |
ENSDART00000040456
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr4_+_13413303 | 0.97 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr7_-_50974318 | 0.97 |
ENSDART00000174297
|
CR354588.1
|
ENSDARG00000105693 |
| chr8_+_13327234 | 0.97 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr15_+_23421623 | 0.96 |
ENSDART00000113279
|
rnf26
|
ring finger protein 26 |
| chr17_+_5776013 | 0.96 |
ENSDART00000168326
|
znf513b
|
zinc finger protein 513b |
| chr20_+_7337986 | 0.96 |
ENSDART00000140222
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr1_-_51863300 | 0.96 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr12_+_27444732 | 0.95 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr20_-_14885722 | 0.95 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr7_+_34235297 | 0.94 |
ENSDART00000173957
|
rfx7
|
regulatory factor X, 7 |
| chr8_+_29885318 | 0.94 |
ENSDART00000154309
|
CR376747.1
|
ENSDARG00000097806 |
| chr8_-_22265875 | 0.94 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr23_+_26153043 | 0.94 |
ENSDART00000054013
|
atp6ap1b
|
ATPase, H+ transporting, lysosomal accessory protein 1b |
| chr1_+_45907424 | 0.94 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr19_+_30212276 | 0.94 |
|
|
|
| chr21_+_33216466 | 0.93 |
ENSDART00000163808
|
ndst1b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1b |
| chr5_-_56641212 | 0.93 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr3_-_42948612 | 0.92 |
|
|
|
| chr16_+_36814839 | 0.91 |
|
|
|
| chr11_-_44756789 | 0.91 |
ENSDART00000161712
|
syngr2b
|
synaptogyrin 2b |
| chr15_+_5125138 | 0.91 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr16_-_12362566 | 0.91 |
ENSDART00000116542
|
U7
|
U7 small nuclear RNA |
| chr7_-_59284425 | 0.91 |
ENSDART00000101731
|
larp7
|
La ribonucleoprotein domain family, member 7 |
| chr2_-_45810714 | 0.91 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr8_-_22266041 | 0.91 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr7_-_54407355 | 0.90 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr5_-_35997345 | 0.90 |
ENSDART00000122098
|
rhogc
|
ras homolog gene family, member Gc |
| chr19_+_17899173 | 0.89 |
ENSDART00000123519
|
ube2e1
|
ubiquitin-conjugating enzyme E2E 1 |
| chr21_+_10692781 | 0.89 |
|
|
|
| chr16_-_2642235 | 0.89 |
|
|
|
| chr22_-_680961 | 0.89 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr6_+_9735100 | 0.89 |
ENSDART00000171518
|
abi2b
|
abl-interactor 2b |
| chr11_+_44916426 | 0.89 |
|
|
|
| chr15_+_6462447 | 0.88 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr12_+_5067663 | 0.87 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr20_-_9212089 | 0.87 |
ENSDART00000064140
|
ylpm1
|
YLP motif containing 1 |
| chr5_+_61389881 | 0.87 |
ENSDART00000141574
|
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr17_-_6443673 | 0.87 |
ENSDART00000129950
ENSDART00000154074 |
ANKRD66
|
si:ch211-189e2.2 |
| chr13_-_37530793 | 0.87 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr3_-_42948574 | 0.87 |
|
|
|
| chr16_+_25097165 | 0.86 |
ENSDART00000157312
|
znf1035
|
zinc finger protein 1035 |
| chr17_+_15527525 | 0.86 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr6_+_135255 | 0.86 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr7_+_56076854 | 0.86 |
|
|
|
| chr12_+_9811557 | 0.86 |
ENSDART00000110458
|
fam117ab
|
family with sequence similarity 117, member Ab |
| chr14_+_20941526 | 0.86 |
ENSDART00000138551
|
smim19
|
small integral membrane protein 19 |
| chr21_+_33216675 | 0.85 |
ENSDART00000163808
|
ndst1b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1b |
| chr19_+_30212200 | 0.85 |
|
|
|
| chr5_+_19363942 | 0.85 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr15_-_161052 | 0.85 |
|
|
|
| chr15_+_14656886 | 0.85 |
ENSDART00000162350
|
FBXO46
|
F-box protein 46 |
| chr20_-_3220475 | 0.84 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr5_+_37496875 | 0.84 |
ENSDART00000135428
|
si:ch211-284e13.5
|
si:ch211-284e13.5 |
| chr24_+_12845339 | 0.84 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr1_-_44892558 | 0.84 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr10_-_17214368 | 0.84 |
|
|
|
| chr7_+_73598715 | 0.84 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr4_+_4825461 | 0.83 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr7_-_73890499 | 0.83 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr24_-_19574944 | 0.83 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr25_+_17593164 | 0.83 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr7_-_73620443 | 0.83 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr17_+_30476515 | 0.82 |
|
|
|
| chr19_+_42661987 | 0.82 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr23_-_27579257 | 0.82 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr25_-_20933295 | 0.82 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr25_-_34599241 | 0.81 |
ENSDART00000156727
|
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr10_-_44634139 | 0.81 |
ENSDART00000010056
ENSDART00000169919 |
ccdc62
|
coiled-coil domain containing 62 |
| chr11_+_2527564 | 0.81 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr10_-_6453139 | 0.81 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr8_-_22266750 | 0.81 |
ENSDART00000100042
ENSDART00000138966 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr17_+_28657998 | 0.81 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr7_-_69883542 | 0.80 |
|
|
|
| chr19_+_30212565 | 0.80 |
|
|
|
| chr6_-_3763940 | 0.80 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr9_+_188559 | 0.80 |
ENSDART00000162764
|
rrp1
|
ribosomal RNA processing 1 |
| chr24_-_20496308 | 0.80 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr18_-_26995562 | 0.80 |
ENSDART00000147823
|
rcn2
|
reticulocalbin 2 |
| chr15_-_33669618 | 0.79 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr3_-_42949189 | 0.79 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr11_+_11136919 | 0.79 |
ENSDART00000026135
|
ly75
|
lymphocyte antigen 75 |
| chr19_+_30212487 | 0.79 |
|
|
|
| chr10_-_44634167 | 0.79 |
ENSDART00000010056
ENSDART00000169919 |
ccdc62
|
coiled-coil domain containing 62 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.0 | GO:1990437 | snRNA 2'-O-methylation(GO:1990437) U6 2'-O-snRNA methylation(GO:1990438) |
| 1.0 | 3.0 | GO:1900144 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.7 | 2.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.7 | 2.0 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.6 | 2.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.6 | 2.8 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.6 | 1.7 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.5 | 4.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.5 | 6.1 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.5 | 2.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.5 | 1.9 | GO:1903449 | male sex determination(GO:0030238) androst-4-ene-3,17-dione biosynthetic process(GO:1903449) |
| 0.5 | 1.8 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.5 | 3.6 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.5 | 1.4 | GO:0044706 | adult heart development(GO:0007512) female pregnancy(GO:0007565) embryo implantation(GO:0007566) multi-multicellular organism process(GO:0044706) apelin receptor signaling pathway(GO:0060183) trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 2.9 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 2.1 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.4 | 1.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.4 | 2.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 1.1 | GO:0039535 | negative regulation of type I interferon production(GO:0032480) cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) cellular response to virus(GO:0098586) |
| 0.4 | 1.8 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.3 | 1.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.3 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.3 | 0.9 | GO:0098751 | bone cell development(GO:0098751) |
| 0.3 | 3.7 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.3 | 3.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 3.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.3 | 2.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.3 | 1.3 | GO:0098921 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.2 | 0.7 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.2 | 1.0 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.2 | 0.7 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 0.6 | GO:0021846 | cell proliferation in forebrain(GO:0021846) centriole elongation(GO:0061511) |
| 0.2 | 0.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.2 | 0.7 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.2 | 1.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.2 | 0.7 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 0.7 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 0.5 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.2 | 2.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 3.0 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.2 | 0.5 | GO:0035552 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.2 | 0.5 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.2 | 0.5 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.2 | 0.8 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.5 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.2 | 4.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 1.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 5.6 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 1.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 2.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) regulation of neutrophil migration(GO:1902622) |
| 0.1 | 0.4 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.1 | 1.5 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.5 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.6 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.7 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.3 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 | 0.4 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 3.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.0 | GO:0097324 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 1.1 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.0 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.4 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.5 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.4 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.3 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 10.5 | GO:0030218 | erythrocyte differentiation(GO:0030218) erythrocyte homeostasis(GO:0034101) |
| 0.1 | 2.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 1.4 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.6 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.1 | 0.8 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.7 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 2.0 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.1 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 3.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 2.0 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.1 | 0.1 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.3 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.1 | 0.2 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 0.2 | GO:0071887 | leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.1 | 1.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 2.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.1 | 1.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.1 | 1.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.3 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 1.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 2.2 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 1.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 3.7 | GO:0006006 | glucose metabolic process(GO:0006006) |
| 0.0 | 1.0 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 4.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 2.8 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.9 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 1.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.4 | GO:0051222 | positive regulation of protein transport(GO:0051222) positive regulation of establishment of protein localization(GO:1904951) |
| 0.0 | 1.3 | GO:0098661 | inorganic anion transmembrane transport(GO:0098661) |
| 0.0 | 0.5 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 1.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 1.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 1.6 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 1.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.1 | GO:0003190 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0018200 | peptidyl-glutamic acid modification(GO:0018200) |
| 0.0 | 0.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.5 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.0 | 0.2 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.5 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.1 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 1.2 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.6 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 1.4 | GO:0007188 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.4 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 4.0 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 2.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.7 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.1 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.5 | 5.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.3 | 1.3 | GO:0031251 | PAN complex(GO:0031251) |
| 0.3 | 1.0 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.3 | 1.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 2.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 1.6 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 0.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 1.9 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 1.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 0.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 0.5 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.7 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 1.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.7 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 8.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 5.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.6 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.7 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.3 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 5.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 4.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.9 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0030880 | RNA polymerase complex(GO:0030880) |
| 0.0 | 3.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 8.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 15.6 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.4 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 10.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.6 | 2.8 | GO:0070551 | endoribonuclease activity, cleaving siRNA-paired mRNA(GO:0070551) |
| 0.5 | 1.9 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.4 | 1.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 1.5 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.4 | 2.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 0.9 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.3 | 2.8 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 1.9 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 0.8 | GO:0004617 | phosphoglycerate dehydrogenase activity(GO:0004617) |
| 0.2 | 0.7 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 1.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.4 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.2 | 1.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 2.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.6 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 0.7 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.2 | 0.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 3.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 1.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 0.5 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.2 | 0.5 | GO:0035515 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.4 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.7 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 2.6 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 2.0 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 4.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 0.2 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 0.7 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.3 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 1.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 2.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 3.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 0.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.6 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 0.6 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 4.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.2 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.5 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.1 | 0.3 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 1.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 17.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 3.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.0 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 3.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 3.8 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.9 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 1.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.9 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.5 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 2.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 6.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.6 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.2 | 2.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 5.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 0.8 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.1 | 2.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 2.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.9 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 1.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 0.7 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.2 | 1.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.0 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.1 | 1.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.8 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.7 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 2.8 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 1.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |