Project
DANIO-CODE
Navigation
Downloads
Results for myog
Z-value: 0.67
Transcription factors associated with myog
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
myog
|
ENSDARG00000009438 | myogenin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| myog | dr10_dc_chr11_-_22438900_22439035 | 0.45 | 8.2e-02 | Click! |
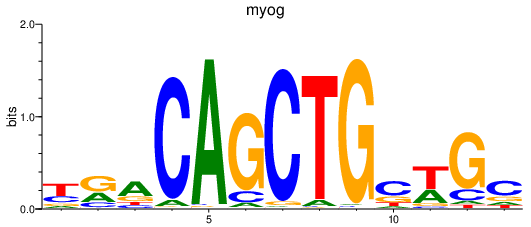
Activity profile of myog motif
Sorted Z-values of myog motif
Network of associatons between targets according to the STRING database.
First level regulatory network of myog
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_27346176 | 1.53 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr3_+_26013873 | 1.32 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr11_-_5868257 | 1.16 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr2_-_21693984 | 1.08 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase-like, a |
| chr24_+_20430778 | 0.92 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr19_+_32317583 | 0.85 |
ENSDART00000151218
|
tpd52
|
tumor protein D52 |
| chr7_+_39173765 | 0.83 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr2_+_42342148 | 0.80 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr6_-_32718634 | 0.79 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr7_+_31567166 | 0.79 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_42006033 | 0.77 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr7_+_6517248 | 0.77 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| KN149932v1_+_27584 | 0.76 |
|
|
|
| chr23_+_22729939 | 0.75 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr3_-_58590651 | 0.73 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr2_-_21677086 | 0.69 |
ENSDART00000057022
|
klhl40a
|
kelch-like family member 40a |
| chr18_+_23198330 | 0.68 |
ENSDART00000143977
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr10_-_28474085 | 0.66 |
|
|
|
| chr24_-_33817169 | 0.65 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr11_+_7148830 | 0.65 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr23_+_43362722 | 0.62 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr20_-_44598129 | 0.60 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr9_-_23081250 | 0.60 |
|
|
|
| chr25_+_35179025 | 0.59 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr19_-_5338129 | 0.58 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr5_-_13318226 | 0.58 |
|
|
|
| chr10_+_34377959 | 0.57 |
|
|
|
| chr22_+_26543146 | 0.56 |
|
|
|
| chr20_+_35479428 | 0.55 |
ENSDART00000159483
ENSDART00000031091 |
BX511024.1
vsnl1a
|
ENSDARG00000104812 visinin-like 1a |
| chr20_+_35479511 | 0.55 |
ENSDART00000135284
|
vsnl1a
|
visinin-like 1a |
| chr4_-_4825948 | 0.55 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr6_-_19432410 | 0.54 |
ENSDART00000006843
|
cacng1a
|
calcium channel, voltage-dependent, gamma subunit 1a |
| chr16_-_42990753 | 0.53 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr11_-_80746 | 0.53 |
|
|
|
| chr8_-_985673 | 0.50 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr25_+_33527870 | 0.49 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr6_+_3771586 | 0.48 |
|
|
|
| chr7_+_40972616 | 0.48 |
ENSDART00000173814
|
scrib
|
scribbled planar cell polarity protein |
| chr25_-_23428527 | 0.48 |
ENSDART00000062930
|
phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr19_+_33235375 | 0.47 |
ENSDART00000052096
|
hrsp12
|
heat-responsive protein 12 |
| chr6_-_42006225 | 0.47 |
ENSDART00000032527
|
cav3
|
caveolin 3 |
| chr5_+_42312784 | 0.46 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr20_-_36984259 | 0.46 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr13_+_30546398 | 0.45 |
|
|
|
| chr10_+_7678087 | 0.45 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr6_+_14823900 | 0.45 |
ENSDART00000178555
|
CU929521.3
|
ENSDARG00000108533 |
| chr16_-_9978112 | 0.45 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr16_-_33637966 | 0.45 |
ENSDART00000142965
|
BX511161.1
|
ENSDARG00000092272 |
| chr21_+_549788 | 0.45 |
ENSDART00000175116
|
CABZ01100207.1
|
ENSDARG00000106163 |
| chr4_-_16365281 | 0.44 |
ENSDART00000139919
|
lum
|
lumican |
| chr9_-_7676791 | 0.44 |
ENSDART00000136438
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr2_-_20462218 | 0.44 |
ENSDART00000124724
|
dpydb
|
dihydropyrimidine dehydrogenase b |
| chr15_+_25554119 | 0.43 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr14_-_30739765 | 0.43 |
ENSDART00000173282
|
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr7_+_49442972 | 0.43 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr18_+_36650892 | 0.42 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr7_-_34656224 | 0.42 |
ENSDART00000073397
|
nfatc3a
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3a |
| chr5_-_63521649 | 0.42 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr7_+_22314304 | 0.41 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr10_+_9716807 | 0.41 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr23_-_18131105 | 0.41 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr3_-_36130248 | 0.41 |
ENSDART00000126588
|
rac3a
|
ras-related C3 botulinum toxin substrate 3a (rho family, small GTP binding protein Rac3) |
| chr3_-_11657881 | 0.41 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr6_+_44817077 | 0.41 |
ENSDART00000169713
|
chl1b
|
cell adhesion molecule L1-like b |
| chr1_-_10822278 | 0.41 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr14_-_47845475 | 0.41 |
ENSDART00000149571
|
ldb2a
|
LIM domain binding 2a |
| chr5_-_47656197 | 0.41 |
|
|
|
| chr16_+_3090170 | 0.41 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr24_-_33817036 | 0.40 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr17_-_24548534 | 0.40 |
|
|
|
| chr10_-_37098396 | 0.40 |
ENSDART00000155277
|
BX323076.2
|
ENSDARG00000097288 |
| chr5_+_71367929 | 0.40 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr5_+_37254520 | 0.39 |
ENSDART00000051222
|
ins
|
preproinsulin |
| chr1_-_24297477 | 0.39 |
|
|
|
| chr14_+_36156947 | 0.39 |
|
|
|
| chr22_-_951880 | 0.39 |
ENSDART00000105895
ENSDART00000172206 |
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr20_-_40422997 | 0.39 |
ENSDART00000075112
|
clvs2
|
clavesin 2 |
| chr9_-_18561215 | 0.39 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr17_+_24421150 | 0.38 |
ENSDART00000168926
|
mdh1ab
|
malate dehydrogenase 1Ab, NAD (soluble) |
| chr10_+_35321228 | 0.38 |
ENSDART00000048831
|
tmem120a
|
transmembrane protein 120A |
| chr14_+_6639864 | 0.38 |
ENSDART00000061001
|
gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr7_-_69399273 | 0.38 |
ENSDART00000159823
ENSDART00000075178 ENSDART00000168942 ENSDART00000126739 |
tspan5a
|
tetraspanin 5a |
| chr20_+_23392782 | 0.38 |
|
|
|
| chr4_-_5589261 | 0.38 |
ENSDART00000017349
|
vegfab
|
vascular endothelial growth factor Ab |
| chr17_-_4160288 | 0.37 |
ENSDART00000153824
|
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr6_-_10728582 | 0.37 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr17_+_8874210 | 0.37 |
|
|
|
| chr1_+_38834751 | 0.36 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr11_-_41357639 | 0.36 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr5_+_41497442 | 0.36 |
ENSDART00000171919
ENSDART00000097587 |
p2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr17_-_35015584 | 0.35 |
ENSDART00000145664
ENSDART00000021128 |
kidins220a
|
kinase D-interacting substrate 220a |
| chr21_-_19789339 | 0.35 |
|
|
|
| chr14_-_8975187 | 0.35 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr4_+_51316598 | 0.35 |
|
|
|
| chr10_-_10372266 | 0.35 |
|
|
|
| chr1_+_36476280 | 0.34 |
ENSDART00000043855
|
dclk2a
|
doublecortin-like kinase 2a |
| chr17_+_52736844 | 0.34 |
ENSDART00000160507
|
meis2a
|
Meis homeobox 2a |
| chr25_-_21618526 | 0.34 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
dedicator of cytokinesis 4 |
| chr18_+_25016753 | 0.34 |
ENSDART00000099476
|
fam174b
|
family with sequence similarity 174, member B |
| chr10_-_38206238 | 0.34 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr17_+_34168095 | 0.34 |
|
|
|
| chr16_-_14463682 | 0.33 |
ENSDART00000011224
|
itga10
|
integrin, alpha 10 |
| chr13_-_10940395 | 0.33 |
ENSDART00000135989
|
cep170aa
|
centrosomal protein 170Aa |
| chr19_+_33235087 | 0.32 |
ENSDART00000052097
ENSDART00000052096 |
hrsp12
|
heat-responsive protein 12 |
| chr13_+_51154848 | 0.32 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr5_+_36180981 | 0.32 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr17_+_27417635 | 0.31 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr18_-_3244618 | 0.31 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr23_-_18131209 | 0.31 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr25_-_20160885 | 0.31 |
ENSDART00000160700
|
dnm1l
|
dynamin 1-like |
| chr3_-_48458042 | 0.31 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr6_-_586251 | 0.31 |
ENSDART00000148867
ENSDART00000149414 ENSDART00000149248 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr5_-_62821458 | 0.31 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr3_+_31821397 | 0.31 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr2_+_27344633 | 0.31 |
ENSDART00000178275
|
cdh7
|
cadherin 7, type 2 |
| chr16_+_5411816 | 0.31 |
|
|
|
| chr14_+_21531709 | 0.30 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr22_+_20695983 | 0.30 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr8_+_44933065 | 0.30 |
ENSDART00000098567
|
ENSDARG00000016545
|
ENSDARG00000016545 |
| chr18_-_1577633 | 0.30 |
|
|
|
| chr14_-_28228583 | 0.30 |
ENSDART00000054088
|
zgc:113364
|
zgc:113364 |
| chr20_+_54525614 | 0.29 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr20_-_54591757 | 0.29 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr19_+_43316957 | 0.29 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr5_+_6233655 | 0.29 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr15_-_10532992 | 0.29 |
ENSDART00000175825
|
tenm4
|
teneurin transmembrane protein 4 |
| chr6_-_16267366 | 0.28 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr12_+_22734974 | 0.28 |
ENSDART00000130594
|
afap1
|
actin filament associated protein 1 |
| chr15_+_29343644 | 0.28 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr20_+_1257719 | 0.28 |
ENSDART00000129777
|
lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr2_+_51265445 | 0.28 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr16_-_9075915 | 0.28 |
|
|
|
| chr14_+_11151485 | 0.28 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr17_+_52736192 | 0.28 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr16_-_14184394 | 0.27 |
ENSDART00000090234
|
trim109
|
tripartite motif containing 109 |
| chr23_+_43284714 | 0.27 |
|
|
|
| chr9_-_43360662 | 0.27 |
ENSDART00000135365
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr19_+_7880087 | 0.27 |
ENSDART00000053380
|
hax1
|
HCLS1 associated protein X-1 |
| chr25_-_21796677 | 0.27 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr2_+_32033028 | 0.27 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr23_+_20050920 | 0.27 |
ENSDART00000144790
|
cfap126
|
cilia and flagella associated protein 126 |
| chr16_+_24032160 | 0.26 |
ENSDART00000103190
|
apoa4b.2
|
apolipoprotein A-IV b, tandem duplicate 2 |
| chr19_-_2239102 | 0.26 |
ENSDART00000024847
|
tmem196a
|
transmembrane protein 196a |
| chr17_+_52736535 | 0.26 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr8_+_44933013 | 0.25 |
ENSDART00000098567
|
ENSDARG00000016545
|
ENSDARG00000016545 |
| chr5_+_56649893 | 0.25 |
ENSDART00000036331
|
atp6v1g1
|
ATPase, H+ transporting, lysosomal, V1 subunit G1 |
| chr18_+_30391910 | 0.25 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr1_+_31210417 | 0.25 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr22_-_28922834 | 0.25 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr25_+_31457309 | 0.24 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr5_-_18895882 | 0.24 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr25_+_33364224 | 0.24 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr12_+_5495284 | 0.24 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr11_+_5761883 | 0.24 |
|
|
|
| chr7_-_72278552 | 0.24 |
ENSDART00000168532
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr10_+_15819127 | 0.24 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr9_+_4410206 | 0.24 |
|
|
|
| KN149895v1_+_73008 | 0.23 |
|
|
|
| chr21_+_45325383 | 0.23 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr8_-_50270783 | 0.23 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr23_-_3466041 | 0.23 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr14_-_41311458 | 0.23 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor-like 1b |
| chr4_+_10065500 | 0.23 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr11_+_41802482 | 0.23 |
ENSDART00000173022
|
camta1
|
calmodulin binding transcription activator 1 |
| chr18_-_47026460 | 0.23 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr25_+_30746445 | 0.23 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr21_+_4964891 | 0.23 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr11_-_11487856 | 0.23 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr17_-_7990960 | 0.23 |
|
|
|
| chr22_-_27274035 | 0.22 |
ENSDART00000153589
|
si:dkey-16m19.1
|
si:dkey-16m19.1 |
| chr10_-_22834248 | 0.22 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr16_-_32628744 | 0.22 |
ENSDART00000137936
|
fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr4_-_5589227 | 0.22 |
ENSDART00000136229
|
vegfab
|
vascular endothelial growth factor Ab |
| chr20_-_49965171 | 0.22 |
|
|
|
| chr2_-_52861662 | 0.22 |
ENSDART00000002241
|
chico
|
chico |
| chr1_-_48853800 | 0.22 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr13_+_781725 | 0.22 |
ENSDART00000129866
|
pcsk2
|
proprotein convertase subtilisin/kexin type 2 |
| chr14_-_16023198 | 0.21 |
|
|
|
| chr12_+_4821529 | 0.21 |
|
|
|
| chr10_-_38206060 | 0.21 |
ENSDART00000174882
|
FP016248.1
|
ENSDARG00000107804 |
| chr15_+_22637181 | 0.21 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr9_-_7676688 | 0.21 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr22_-_29387056 | 0.21 |
ENSDART00000121599
|
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr15_-_46484516 | 0.21 |
ENSDART00000154577
|
zgc:162872
|
zgc:162872 |
| chr12_+_41016238 | 0.21 |
ENSDART00000170976
|
kif5bb
|
kinesin family member 5B, b |
| chr1_-_18809429 | 0.21 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr7_-_6226607 | 0.21 |
ENSDART00000129239
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr14_-_32877752 | 0.20 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr19_+_38033219 | 0.20 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr19_+_177005 | 0.20 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr24_-_28762107 | 0.20 |
|
|
|
| chr3_+_56970554 | 0.20 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr4_+_8531280 | 0.20 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr2_+_32033176 | 0.20 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr19_-_9793494 | 0.20 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr20_+_34248780 | 0.20 |
ENSDART00000152870
|
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr10_-_34927807 | 0.19 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr17_-_1496133 | 0.19 |
ENSDART00000175509
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr4_+_16798009 | 0.19 |
ENSDART00000039027
|
golt1ba
|
golgi transport 1Ba |
| chr5_+_60201330 | 0.19 |
ENSDART00000009298
|
tmem248
|
transmembrane protein 248 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 | 1.3 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 0.4 | 1.2 | GO:1903060 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.4 | 1.2 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.8 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.6 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.5 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 1.4 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.4 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.4 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.4 | GO:0019859 | thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.3 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.6 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.4 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of feeding behavior(GO:0060259) |
| 0.1 | 0.8 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.1 | 0.7 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 0.3 | GO:2000543 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) positive regulation of gastrulation(GO:2000543) |
| 0.1 | 1.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.4 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.1 | 0.5 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.1 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 0.5 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.4 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.1 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.4 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.2 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.2 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.0 | 0.8 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.2 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.1 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.4 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 1.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.5 | GO:0009261 | purine ribonucleotide catabolic process(GO:0009154) ribonucleotide catabolic process(GO:0009261) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.4 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.1 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.0 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.7 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 1.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 1.2 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.1 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 0.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.4 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.5 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.3 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.2 | 0.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.1 | 1.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.4 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0016615 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.5 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.4 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.6 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.0 | 0.1 | GO:0019871 | voltage-gated sodium channel activity(GO:0005248) sodium channel inhibitor activity(GO:0019871) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0019870 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |