Project
DANIO-CODE
Navigation
Downloads
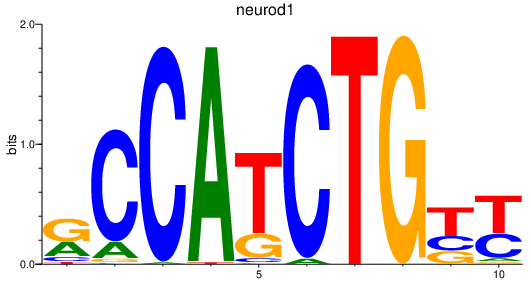
Results for neurod1
Z-value: 0.81
Transcription factors associated with neurod1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
neurod1
|
ENSDARG00000019566 | neuronal differentiation 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| neurod1 | dr10_dc_chr9_-_44493074_44493168 | 0.79 | 2.8e-04 | Click! |
Activity profile of neurod1 motif
Sorted Z-values of neurod1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of neurod1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_55204516 | 2.08 |
|
|
|
| chr22_+_26543146 | 1.91 |
|
|
|
| chr5_-_36237656 | 1.68 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr5_-_28090077 | 1.67 |
|
|
|
| chr9_-_7695437 | 1.64 |
ENSDART00000102715
|
tuba8l3
|
tubulin, alpha 8 like 3 |
| chr9_+_308260 | 1.57 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr10_+_15819127 | 1.49 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr21_+_27346176 | 1.49 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr2_-_21693984 | 1.32 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase-like, a |
| chr1_+_51189343 | 1.28 |
ENSDART00000150757
|
BX548032.3
|
ENSDARG00000096092 |
| chr11_+_24687813 | 1.27 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr18_+_30391910 | 1.26 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr3_-_48865474 | 1.24 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr7_+_32451041 | 1.23 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr22_+_12327849 | 1.20 |
ENSDART00000178678
|
FO704572.1
|
ENSDARG00000106890 |
| chr9_-_44493074 | 1.18 |
ENSDART00000167685
|
neurod1
|
neuronal differentiation 1 |
| chr16_+_46327528 | 1.18 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr6_-_15526547 | 1.17 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr17_+_27383737 | 1.16 |
ENSDART00000156756
|
FP017274.1
|
ENSDARG00000097369 |
| chr10_-_42032702 | 1.15 |
|
|
|
| chr10_-_43826919 | 1.15 |
ENSDART00000039551
ENSDART00000160786 ENSDART00000099134 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr1_+_51925783 | 1.14 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr1_-_28241500 | 1.13 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr8_+_7340538 | 1.13 |
ENSDART00000121708
|
pcsk1nl
|
proprotein convertase subtilisin/kexin type 1 inhibitor, like |
| chr5_+_36093701 | 1.12 |
ENSDART00000019259
|
dlb
|
deltaB |
| chr15_+_29153215 | 1.09 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr21_-_23710216 | 1.09 |
ENSDART00000132570
|
ncam1a
|
neural cell adhesion molecule 1a |
| chr18_+_20504980 | 1.08 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr16_+_46034246 | 1.08 |
ENSDART00000101753
|
mtmr11
|
myotubularin related protein 11 |
| chr5_-_40133824 | 1.05 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr6_-_43094573 | 1.05 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr2_+_2895642 | 1.03 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr15_+_7179382 | 1.02 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr10_+_4717800 | 1.01 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr16_+_50189177 | 1.01 |
ENSDART00000163565
|
plcl2
|
phospholipase C-like 2 |
| chr4_-_4423059 | 1.00 |
ENSDART00000082046
|
ntf3
|
neurotrophin 3 |
| chr16_-_10404109 | 0.99 |
|
|
|
| chr12_-_41124276 | 0.99 |
ENSDART00000172022
ENSDART00000158605 ENSDART00000162967 |
dpysl4
|
dihydropyrimidinase-like 4 |
| chr3_-_28120668 | 0.98 |
|
|
|
| chr22_+_34738389 | 0.98 |
ENSDART00000154372
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr23_+_35964754 | 0.97 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr19_+_34145030 | 0.97 |
ENSDART00000151521
|
runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr5_-_47656197 | 0.95 |
|
|
|
| chr24_+_7607915 | 0.95 |
ENSDART00000124409
|
cavin1b
|
caveolae associated protein 1b |
| chr2_-_30675594 | 0.95 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr10_+_15819085 | 0.95 |
ENSDART00000114483
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr6_-_9964616 | 0.95 |
|
|
|
| chr5_-_51172369 | 0.94 |
ENSDART00000164267
ENSDART00000157866 |
homer1b
|
homer scaffolding protein 1b |
| chr16_-_37461878 | 0.94 |
ENSDART00000142916
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr23_+_21546553 | 0.93 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr7_+_28862183 | 0.92 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr8_-_11191783 | 0.92 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr10_+_35344246 | 0.92 |
ENSDART00000139229
|
pora
|
P450 (cytochrome) oxidoreductase a |
| chr13_+_25319230 | 0.91 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr18_+_23198330 | 0.91 |
ENSDART00000143977
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr19_-_34328673 | 0.90 |
ENSDART00000164563
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr6_+_27156169 | 0.89 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr19_+_48499602 | 0.89 |
|
|
|
| chr6_+_56157608 | 0.89 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr5_+_42312784 | 0.87 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr16_+_24057260 | 0.86 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr23_+_35609887 | 0.85 |
ENSDART00000179393
|
tuba1b
|
tubulin, alpha 1b |
| chr2_+_25363717 | 0.85 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr8_-_985673 | 0.84 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr1_-_37990863 | 0.83 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr13_+_28723589 | 0.83 |
|
|
|
| chr6_-_6835861 | 0.83 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr13_+_25298383 | 0.83 |
|
|
|
| chr7_+_6517248 | 0.83 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr23_-_1008307 | 0.82 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr6_-_9329917 | 0.82 |
ENSDART00000151470
|
map3k2
|
mitogen-activated protein kinase kinase kinase 2 |
| chr7_-_55406347 | 0.82 |
ENSDART00000021009
|
cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr24_+_24924379 | 0.81 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr21_-_39239757 | 0.78 |
|
|
|
| chr5_+_37254520 | 0.78 |
ENSDART00000051222
|
ins
|
preproinsulin |
| chr9_-_1958795 | 0.77 |
ENSDART00000138730
|
hoxd3a
|
homeobox D3a |
| chr1_+_9999665 | 0.76 |
ENSDART00000054879
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr5_-_45729685 | 0.76 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr25_-_8475902 | 0.76 |
ENSDART00000176751
|
CU929451.3
|
ENSDARG00000107886 |
| chr24_-_34449257 | 0.75 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr13_-_16126849 | 0.75 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr8_-_50159285 | 0.75 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr16_+_24717056 | 0.75 |
ENSDART00000157237
|
si:dkey-56f14.7
|
si:dkey-56f14.7 |
| chr5_-_33169062 | 0.74 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr2_-_42279180 | 0.74 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr25_-_28932291 | 0.74 |
ENSDART00000073488
|
CU469568.1
|
ENSDARG00000051805 |
| chr23_-_10242377 | 0.73 |
ENSDART00000129044
|
krt5
|
keratin 5 |
| chr6_-_11544518 | 0.72 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr23_+_21610798 | 0.72 |
ENSDART00000111966
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr5_+_40722565 | 0.71 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr11_-_32461160 | 0.71 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr5_+_29327479 | 0.71 |
|
|
|
| chr8_-_23744125 | 0.70 |
ENSDART00000141871
|
INAVA
|
innate immunity activator |
| chr12_+_30448812 | 0.69 |
ENSDART00000126064
|
si:ch211-28p3.4
|
si:ch211-28p3.4 |
| chr6_-_60088120 | 0.69 |
|
|
|
| chr25_-_26292712 | 0.68 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr15_+_1832874 | 0.68 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr15_+_3296905 | 0.67 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr3_+_17210396 | 0.67 |
ENSDART00000090676
|
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr17_-_52735250 | 0.66 |
|
|
|
| chr9_+_23854967 | 0.66 |
ENSDART00000060905
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr10_+_33956052 | 0.66 |
|
|
|
| chr15_+_15580544 | 0.66 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr2_+_50892294 | 0.65 |
ENSDART00000018150
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr3_-_1435654 | 0.65 |
ENSDART00000089646
ENSDART00000158110 |
fam234b
|
family with sequence similarity 234, member B |
| chr10_-_30016761 | 0.64 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr10_-_15896595 | 0.64 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr11_-_23206021 | 0.64 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr16_-_32059848 | 0.64 |
ENSDART00000138701
|
gstk1
|
glutathione S-transferase kappa 1 |
| chr19_-_27676873 | 0.64 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr8_-_7425677 | 0.63 |
ENSDART00000149671
|
hdac6
|
histone deacetylase 6 |
| chr6_-_43094926 | 0.63 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr1_-_40587797 | 0.62 |
ENSDART00000170895
|
rgs12b
|
regulator of G protein signaling 12b |
| chr5_+_23741791 | 0.62 |
ENSDART00000049003
|
atp6v1aa
|
ATPase, H+ transporting, lysosomal, V1 subunit Aa |
| chr14_+_2969679 | 0.60 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr3_+_49277125 | 0.60 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr24_+_35173816 | 0.60 |
|
|
|
| chr5_+_29327363 | 0.59 |
|
|
|
| chr4_-_9591451 | 0.59 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr15_-_4537178 | 0.58 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr18_-_47026460 | 0.58 |
ENSDART00000108574
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr13_-_24181106 | 0.58 |
ENSDART00000004420
|
rab4a
|
RAB4a, member RAS oncogene family |
| chr11_+_39768718 | 0.57 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr16_+_5625301 | 0.57 |
|
|
|
| chr16_-_33976031 | 0.57 |
ENSDART00000110743
|
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr25_+_34515221 | 0.57 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr25_+_7391458 | 0.57 |
ENSDART00000042928
|
fuk
|
fucokinase |
| chr10_-_28474085 | 0.57 |
|
|
|
| chr7_+_72128846 | 0.57 |
ENSDART00000123887
|
MAPK8IP1 (1 of many)
|
mitogen-activated protein kinase 8 interacting protein 1 |
| chr21_+_32285006 | 0.57 |
|
|
|
| chr24_-_38758093 | 0.56 |
|
|
|
| chr1_+_52308366 | 0.56 |
ENSDART00000108492
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr15_+_19358079 | 0.56 |
ENSDART00000123815
|
jam3a
|
junctional adhesion molecule 3a |
| chr17_-_37040941 | 0.55 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr11_-_25803101 | 0.55 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr1_-_48853800 | 0.55 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr8_-_14113377 | 0.55 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr20_+_35377079 | 0.54 |
ENSDART00000168216
|
fam49a
|
family with sequence similarity 49, member A |
| chr23_-_19053587 | 0.53 |
|
|
|
| chr8_+_28513670 | 0.53 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr10_-_13220838 | 0.53 |
ENSDART00000160265
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr22_+_24131239 | 0.52 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr23_-_15381030 | 0.52 |
|
|
|
| chr8_-_16556749 | 0.52 |
ENSDART00000101655
|
calr
|
calreticulin |
| chr19_-_27676807 | 0.52 |
ENSDART00000045616
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr20_+_23599157 | 0.52 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr23_-_18131105 | 0.52 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr7_-_68629353 | 0.52 |
|
|
|
| chr20_-_49965171 | 0.52 |
|
|
|
| chr23_-_15807415 | 0.52 |
ENSDART00000178557
|
BX005011.3
|
ENSDARG00000108083 |
| chr12_+_22137783 | 0.51 |
ENSDART00000131175
|
wnt3
|
wingless-type MMTV integration site family, member 3 |
| chr6_+_30441419 | 0.50 |
|
|
|
| chr18_+_36825511 | 0.50 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr11_+_36781811 | 0.50 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr5_+_3172101 | 0.50 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr22_-_17026124 | 0.50 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr7_-_14913632 | 0.49 |
|
|
|
| chr8_-_6899611 | 0.49 |
ENSDART00000139545
|
wdr13
|
WD repeat domain 13 |
| chr23_+_37535830 | 0.49 |
|
|
|
| chr16_+_50189117 | 0.49 |
ENSDART00000163565
|
plcl2
|
phospholipase C-like 2 |
| chr6_+_54134129 | 0.49 |
ENSDART00000156554
|
hmga1b
|
high mobility group AT-hook 1b |
| chr16_-_51287698 | 0.48 |
ENSDART00000156255
|
ago1
|
argonaute RISC catalytic component 1 |
| chr3_+_26023021 | 0.48 |
|
|
|
| chr2_-_30005139 | 0.48 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr1_-_18809429 | 0.48 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr1_+_9999842 | 0.48 |
ENSDART00000152438
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr20_-_49965261 | 0.48 |
|
|
|
| chr18_-_14868444 | 0.48 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr5_-_62116975 | 0.47 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr14_+_6117282 | 0.47 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr25_-_35539585 | 0.46 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr3_-_55995924 | 0.46 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr19_+_35415174 | 0.46 |
ENSDART00000103266
|
wdyhv1
|
WDYHV motif containing 1 |
| chr17_-_52493315 | 0.46 |
ENSDART00000156806
|
rps6kl1
|
ribosomal protein S6 kinase-like 1 |
| chr3_+_33209227 | 0.46 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr7_-_29070193 | 0.46 |
ENSDART00000147251
|
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr7_+_20215723 | 0.46 |
ENSDART00000173724
ENSDART00000173773 |
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr12_-_3903961 | 0.45 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr18_+_12179227 | 0.45 |
ENSDART00000162067
ENSDART00000168386 |
fgd4a
|
FYVE, RhoGEF and PH domain containing 4a |
| chr18_+_15827187 | 0.45 |
ENSDART00000147024
ENSDART00000099944 ENSDART00000160470 |
nudt4a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4a |
| chr10_+_9134634 | 0.44 |
ENSDART00000110443
|
fstb
|
follistatin b |
| chr14_-_136402 | 0.44 |
|
|
|
| chr1_+_11400375 | 0.44 |
|
|
|
| chr10_-_17383466 | 0.44 |
|
|
|
| chr1_-_55162455 | 0.44 |
ENSDART00000137909
|
samd1b
|
sterile alpha motif domain containing 1b |
| chr3_-_28120760 | 0.44 |
|
|
|
| chr5_+_43365449 | 0.44 |
ENSDART00000113502
|
TMEM8B
|
si:dkey-84j12.1 |
| chr3_+_38651644 | 0.44 |
ENSDART00000083394
|
si:dkey-106c17.3
|
si:dkey-106c17.3 |
| chr5_+_29327447 | 0.44 |
|
|
|
| chr16_+_5625552 | 0.43 |
|
|
|
| chr10_-_24401876 | 0.43 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr25_+_16593169 | 0.43 |
ENSDART00000073416
|
cecr1a
|
cat eye syndrome chromosome region, candidate 1a |
| chr9_+_33177527 | 0.43 |
ENSDART00000174533
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr25_-_35159840 | 0.43 |
ENSDART00000148718
ENSDART00000164189 ENSDART00000027174 |
lrrk2
|
leucine-rich repeat kinase 2 |
| chr3_+_17760847 | 0.42 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr1_+_2074153 | 0.42 |
ENSDART00000058877
|
rap2ab
|
RAP2A, member of RAS oncogene family b |
| chr14_-_24304373 | 0.42 |
|
|
|
| chr12_-_36255806 | 0.42 |
ENSDART00000159513
|
si:dkey-113d16.9
|
si:dkey-113d16.9 |
| chr19_-_33301572 | 0.42 |
ENSDART00000052080
|
laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr22_+_13862110 | 0.42 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.4 | 1.6 | GO:0070296 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.4 | 1.1 | GO:0098810 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.4 | 1.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.3 | 1.0 | GO:0003091 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) carbon dioxide transport(GO:0015670) |
| 0.3 | 2.0 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.3 | 0.9 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.3 | 1.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.2 | 1.7 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 0.7 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.2 | 0.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 1.7 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.2 | 0.8 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of feeding behavior(GO:0060259) |
| 0.2 | 0.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 2.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 0.3 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.2 | 1.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 1.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.2 | 0.9 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.1 | 0.7 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 0.4 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.5 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.1 | 1.2 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.1 | 0.8 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 1.4 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 1.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.4 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.1 | 0.7 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.1 | 0.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.3 | GO:0060296 | regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.1 | 0.3 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 0.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.7 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.4 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 1.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.6 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0048852 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.9 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.9 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.9 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.8 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.3 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.0 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.5 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.6 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.6 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 2.0 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.9 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.9 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 1.1 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.8 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 2.0 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 1.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 2.4 | GO:0098916 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.5 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.6 | GO:0006476 | protein deacetylation(GO:0006476) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 1.2 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.7 | GO:0031672 | A band(GO:0031672) |
| 0.1 | 0.3 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.4 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 1.9 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 2.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 1.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0099738 | basal cortex(GO:0045180) cell cortex region(GO:0099738) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.3 | 1.0 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.3 | 1.6 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 1.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.2 | 1.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 0.6 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.2 | 0.8 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.9 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 0.9 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 2.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 1.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.8 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.8 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.9 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.1 | 0.9 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.2 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.4 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 3.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.5 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.4 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 2.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.7 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.3 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.5 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.5 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 0.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |