Project
DANIO-CODE
Navigation
Downloads
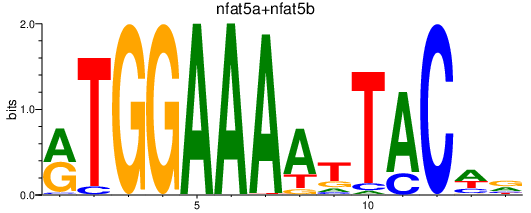
Results for nfat5a+nfat5b
Z-value: 0.74
Transcription factors associated with nfat5a+nfat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfat5a
|
ENSDARG00000020872 | nuclear factor of activated T cells 5a |
|
nfat5b
|
ENSDARG00000102556 | nuclear factor of activated T cells 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfat5b | dr10_dc_chr7_+_67102061_67102089 | 0.66 | 5.7e-03 | Click! |
| nfat5a | dr10_dc_chr25_-_36550392_36550516 | 0.31 | 2.5e-01 | Click! |
Activity profile of nfat5a+nfat5b motif
Sorted Z-values of nfat5a+nfat5b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfat5a+nfat5b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_30013086 | 1.24 |
ENSDART00000138050
|
rbm33b
|
RNA binding motif protein 33b |
| chr23_-_31719203 | 1.23 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr14_-_34293596 | 1.12 |
ENSDART00000128869
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr15_-_25164321 | 1.10 |
ENSDART00000154628
|
fam101b
|
family with sequence similarity 101, member B |
| chr21_-_32747592 | 1.04 |
|
|
|
| chr4_+_75515863 | 0.94 |
ENSDART00000163676
|
CU467646.6
|
ENSDARG00000104678 |
| chr7_-_2614300 | 0.94 |
ENSDART00000153548
|
BX323987.1
|
ENSDARG00000097745 |
| chr22_+_4035577 | 0.93 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr21_+_21242470 | 0.93 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr2_-_6203659 | 0.92 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr3_+_59972297 | 0.92 |
|
|
|
| chr10_+_15296880 | 0.90 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr24_-_19574944 | 0.79 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr23_+_2786407 | 0.76 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr13_+_28785104 | 0.72 |
|
|
|
| chr2_-_22631167 | 0.70 |
ENSDART00000168653
ENSDART00000158558 |
fam110b
BX927330.1
|
family with sequence similarity 110, member B ENSDARG00000098356 |
| chr3_+_31948579 | 0.69 |
ENSDART00000124943
|
aldh16a1
|
aldehyde dehydrogenase 16 family, member A1 |
| chr1_-_44893257 | 0.68 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_+_30266913 | 0.67 |
ENSDART00000109729
|
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr21_-_32747544 | 0.66 |
|
|
|
| chr22_+_1663690 | 0.65 |
ENSDART00000166672
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr12_+_30389706 | 0.65 |
|
|
|
| chr7_+_31750514 | 0.64 |
ENSDART00000173976
|
mettl15
|
methyltransferase like 15 |
| chr9_+_33334773 | 0.63 |
ENSDART00000005879
|
atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr16_-_21241981 | 0.62 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr14_-_30604897 | 0.61 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr20_-_22631007 | 0.60 |
|
|
|
| chr5_-_56641212 | 0.59 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr1_-_53052062 | 0.58 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr15_-_18273640 | 0.56 |
ENSDART00000141508
|
btr16
|
bloodthirsty-related gene family, member 16 |
| chr19_-_25564720 | 0.54 |
ENSDART00000148432
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr9_+_34510091 | 0.53 |
ENSDART00000035522
ENSDART00000170615 ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr1_-_7882397 | 0.51 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr22_-_8276968 | 0.51 |
ENSDART00000123982
|
CABZ01077218.1
|
ENSDARG00000089720 |
| chr7_-_56472076 | 0.50 |
|
|
|
| chr12_-_17481026 | 0.49 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr21_+_31216418 | 0.48 |
ENSDART00000178521
|
asl
|
argininosuccinate lyase |
| chr6_-_22910576 | 0.48 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr9_-_32240567 | 0.47 |
|
|
|
| chr3_-_32457708 | 0.46 |
|
|
|
| chr7_+_28844711 | 0.45 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr25_-_34631252 | 0.44 |
ENSDART00000109751
|
zgc:173585
|
zgc:173585 |
| chr24_-_19574666 | 0.42 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr6_-_12665211 | 0.41 |
ENSDART00000150887
|
ical1
|
islet cell autoantigen 1-like |
| chr22_-_20394671 | 0.40 |
ENSDART00000105520
|
pias4a
|
protein inhibitor of activated STAT, 4a |
| chr24_+_41409608 | 0.39 |
ENSDART00000176003
|
nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr7_-_31749636 | 0.38 |
ENSDART00000023568
|
kif18a
|
kinesin family member 18A |
| chr4_+_10605257 | 0.38 |
ENSDART00000122636
ENSDART00000037140 |
ing3
|
inhibitor of growth family, member 3 |
| chr17_+_43022709 | 0.35 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr19_+_3115685 | 0.35 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_40387858 | 0.35 |
ENSDART00000055194
|
actb2
|
actin, beta 2 |
| chr5_+_1461715 | 0.35 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr9_+_6277952 | 0.34 |
ENSDART00000078525
|
uxs1
|
UDP-glucuronate decarboxylase 1 |
| chr7_+_55648832 | 0.34 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr5_-_9443859 | 0.34 |
|
|
|
| chr19_-_8829701 | 0.34 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr14_+_23673635 | 0.33 |
ENSDART00000124944
|
kif3a
|
kinesin family member 3A |
| chr2_-_30012325 | 0.32 |
ENSDART00000020792
|
cnpy1
|
canopy1 |
| chr24_-_19574761 | 0.32 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr19_+_41596472 | 0.30 |
|
|
|
| chr3_+_21928413 | 0.29 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr3_-_35425227 | 0.29 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr8_+_19472074 | 0.28 |
ENSDART00000158859
ENSDART00000157697 |
sec22bb
|
SEC22 homolog B, vesicle trafficking protein b |
| chr22_-_26231695 | 0.27 |
ENSDART00000142821
|
ccdc130
|
coiled-coil domain containing 130 |
| chr11_-_12024136 | 0.27 |
ENSDART00000111919
|
sp2
|
sp2 transcription factor |
| chr6_-_22910388 | 0.26 |
ENSDART00000164114
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr8_-_12631385 | 0.25 |
ENSDART00000143155
|
CU639468.1
|
ENSDARG00000093851 |
| chr11_-_30364563 | 0.25 |
ENSDART00000165501
|
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr13_-_49826526 | 0.25 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr3_-_5157662 | 0.25 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr7_-_48394268 | 0.23 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr17_+_12504341 | 0.23 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr19_+_33220009 | 0.23 |
ENSDART00000008698
|
stk3
|
serine/threonine kinase 3 (STE20 homolog, yeast) |
| chr12_-_17480978 | 0.22 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr23_-_3760604 | 0.22 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr8_-_4732671 | 0.21 |
ENSDART00000082256
|
ciao1
|
cytosolic iron-sulfur assembly component 1 |
| chr7_+_71766373 | 0.21 |
ENSDART00000011303
|
ywhae2
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 2 |
| chr5_-_12853593 | 0.20 |
ENSDART00000099602
|
ube2l3b
|
ubiquitin-conjugating enzyme E2L 3b |
| chr5_+_41658782 | 0.20 |
ENSDART00000143738
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr7_+_24983985 | 0.18 |
|
|
|
| chr1_+_18610282 | 0.18 |
ENSDART00000129970
|
si:dkeyp-118a3.2
|
si:dkeyp-118a3.2 |
| chr1_-_44874299 | 0.17 |
|
|
|
| chr16_+_43174128 | 0.17 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr24_-_28150352 | 0.16 |
ENSDART00000145290
|
bcl2a
|
B-cell CLL/lymphoma 2a |
| chr9_-_46614949 | 0.16 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr12_-_10403738 | 0.16 |
ENSDART00000014379
|
uqcrc2b
|
ubiquinol-cytochrome c reductase core protein IIb |
| chr2_+_7127672 | 0.15 |
ENSDART00000050597
|
xpr1b
|
xenotropic and polytropic retrovirus receptor 1b |
| chr18_+_16726807 | 0.15 |
ENSDART00000166849
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr2_-_42643265 | 0.14 |
ENSDART00000009093
|
esyt2a
|
extended synaptotagmin-like protein 2a |
| chr18_+_5637539 | 0.14 |
|
|
|
| chr1_+_44248486 | 0.14 |
ENSDART00000067621
|
vps37c
|
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
| chr19_-_10295398 | 0.14 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr3_+_34063993 | 0.13 |
|
|
|
| chr13_+_21973848 | 0.13 |
ENSDART00000115393
|
si:dkey-174i8.1
|
si:dkey-174i8.1 |
| chr11_-_30364626 | 0.12 |
ENSDART00000089803
ENSDART00000046800 ENSDART00000172244 |
slc8a1a
|
solute carrier family 8 (sodium/calcium exchanger), member 1a |
| chr16_-_42857240 | 0.12 |
|
|
|
| chr24_+_36478896 | 0.12 |
|
|
|
| chr24_-_21529774 | 0.12 |
ENSDART00000123216
|
lnx2a
|
ligand of numb-protein X 2a |
| chr25_-_12236333 | 0.11 |
ENSDART00000174863
ENSDART00000179042 |
BX323452.1
|
ENSDARG00000107774 |
| chr20_+_411845 | 0.11 |
ENSDART00000108872
|
ndufaf4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr4_-_12837406 | 0.11 |
|
|
|
| chr18_+_5637588 | 0.10 |
|
|
|
| chr6_-_36574848 | 0.10 |
ENSDART00000135413
|
her6
|
hairy-related 6 |
| chr22_-_31602823 | 0.10 |
|
|
|
| chr14_+_23673541 | 0.09 |
ENSDART00000124944
|
kif3a
|
kinesin family member 3A |
| chr17_+_43022617 | 0.09 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr13_-_16127236 | 0.08 |
ENSDART00000079745
|
zgc:110045
|
zgc:110045 |
| chr3_-_32457740 | 0.07 |
|
|
|
| chr16_-_28901882 | 0.07 |
ENSDART00000142920
|
zgc:162150
|
zgc:162150 |
| chr17_-_23947646 | 0.07 |
|
|
|
| chr9_+_54185729 | 0.07 |
ENSDART00000165991
|
lect1
|
leukocyte cell derived chemotaxin 1 |
| chr19_-_3115104 | 0.07 |
ENSDART00000093146
|
bop1
|
block of proliferation 1 |
| chr7_-_29003965 | 0.06 |
|
|
|
| chr23_-_36023579 | 0.06 |
|
|
|
| chr19_-_10295122 | 0.06 |
ENSDART00000148225
|
znf865
|
zinc finger protein 865 |
| chr8_+_31863748 | 0.06 |
ENSDART00000146933
|
c7a
|
complement component 7a |
| chr17_-_33462243 | 0.06 |
ENSDART00000131807
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr19_-_42847112 | 0.05 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr7_+_39173765 | 0.04 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr7_+_39353662 | 0.04 |
ENSDART00000173847
ENSDART00000173845 |
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr16_+_21122024 | 0.04 |
|
|
|
| chr8_+_50994889 | 0.04 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr7_-_56493164 | 0.03 |
|
|
|
| chr18_+_16726839 | 0.03 |
ENSDART00000166849
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr9_-_11291927 | 0.03 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr5_-_12213712 | 0.03 |
ENSDART00000099749
|
CLDN22
|
claudin 22 |
| chr13_+_6990132 | 0.03 |
ENSDART00000140828
|
CU179758.1
|
ENSDARG00000094815 |
| chr7_-_45747112 | 0.03 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr19_+_3115791 | 0.02 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr18_-_1006445 | 0.02 |
ENSDART00000062654
|
parp6a
|
poly (ADP-ribose) polymerase family, member 6a |
| chr13_-_42630594 | 0.02 |
ENSDART00000111255
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr7_-_28844560 | 0.02 |
ENSDART00000076386
|
fbxl8
|
F-box and leucine-rich repeat protein 8 |
| chr2_-_41669059 | 0.01 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr15_+_11809276 | 0.01 |
ENSDART00000161930
|
strn4
|
striatin, calmodulin binding protein 4 |
| chr2_+_24135188 | 0.01 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr2_-_41669008 | 0.01 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr9_-_46615155 | 0.00 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.4 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.1 | 0.9 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.4 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 0.6 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.1 | 0.5 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.1 | 0.6 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.0 | 0.1 | GO:0030002 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.5 | GO:0009084 | glutamine family amino acid biosynthetic process(GO:0009084) |
| 0.0 | 0.6 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.1 | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.2 | GO:1900006 | membrane tubulation(GO:0097320) positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.9 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.6 | GO:0071424 | rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) |
| 0.1 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.9 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.4 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |