Project
DANIO-CODE
Navigation
Downloads
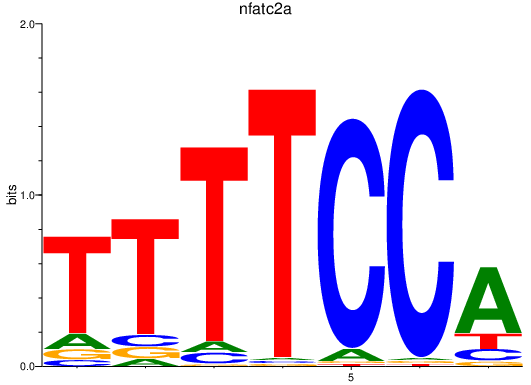
Results for nfatc2a
Z-value: 3.16
Transcription factors associated with nfatc2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc2a
|
ENSDARG00000100927 | nuclear factor of activated T cells 2a |
Activity profile of nfatc2a motif
Sorted Z-values of nfatc2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfatc2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_39315024 | 10.82 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr24_-_28814066 | 7.99 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr16_-_45943282 | 7.41 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr20_+_12559243 | 7.13 |
ENSDART00000172370
|
si:dkey-269i1.4
|
si:dkey-269i1.4 |
| chr17_+_8166167 | 6.81 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr20_-_49880744 | 6.38 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr3_+_24067387 | 6.17 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr4_+_2215426 | 5.99 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr5_+_28559915 | 5.86 |
ENSDART00000088827
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr5_-_30324182 | 5.85 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr22_-_26333957 | 5.73 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr1_-_50831155 | 5.52 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr19_-_41482388 | 5.43 |
ENSDART00000111982
|
sgce
|
sarcoglycan, epsilon |
| chr4_+_7668939 | 5.31 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr4_+_10065500 | 4.95 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr16_+_23482744 | 4.95 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr21_-_5852663 | 4.73 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr19_+_9054095 | 4.57 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr2_-_34010299 | 4.55 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr18_+_38773971 | 4.43 |
ENSDART00000010177
|
onecut1
|
one cut homeobox 1 |
| chr1_-_674449 | 4.38 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr14_-_30363703 | 4.34 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin-like extracellular matrix protein 2a |
| chr22_+_31089813 | 4.34 |
ENSDART00000060005
|
rpl32
|
ribosomal protein L32 |
| chr7_+_21455138 | 4.34 |
ENSDART00000173992
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr23_+_36002332 | 4.32 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr13_-_30515486 | 4.28 |
ENSDART00000139607
|
zcchc24
|
zinc finger, CCHC domain containing 24 |
| chr13_+_22545318 | 4.21 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr8_+_46651079 | 4.20 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr9_+_30279907 | 4.20 |
ENSDART00000102981
|
col8a1a
|
collagen, type VIII, alpha 1a |
| chr3_+_26013873 | 4.19 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr6_-_47842137 | 4.16 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr7_-_54407681 | 4.13 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr11_-_38843219 | 4.11 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr16_-_14506846 | 4.06 |
ENSDART00000170957
|
crabp2a
|
cellular retinoic acid binding protein 2, a |
| chr12_-_3098744 | 4.00 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr23_+_32573474 | 3.96 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr21_+_5528438 | 3.95 |
ENSDART00000168158
|
shroom3
|
shroom family member 3 |
| chr22_-_35967525 | 3.93 |
ENSDART00000176971
|
CABZ01037120.1
|
ENSDARG00000106725 |
| chr9_-_9693163 | 3.87 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr9_-_13383818 | 3.86 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr11_-_26428979 | 3.83 |
ENSDART00000111539
|
efcc1
|
EF-hand and coiled-coil domain containing 1 |
| chr4_+_1667305 | 3.82 |
ENSDART00000150198
|
slc38a4
|
solute carrier family 38, member 4 |
| chr1_-_53746151 | 3.81 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr6_-_49527428 | 3.73 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr3_-_23444371 | 3.69 |
ENSDART00000087726
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr15_+_34131126 | 3.68 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr10_+_4717800 | 3.64 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr8_-_38357549 | 3.62 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr7_+_38479571 | 3.62 |
ENSDART00000170486
|
f2
|
coagulation factor II (thrombin) |
| chr16_+_29558320 | 3.61 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr15_+_125263 | 3.54 |
|
|
|
| chr6_-_48095609 | 3.51 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr5_-_58162552 | 3.49 |
ENSDART00000161230
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr24_-_9151388 | 3.49 |
ENSDART00000149875
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_-_21265803 | 3.44 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr18_+_40364675 | 3.43 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr24_-_33869817 | 3.40 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr2_-_43998886 | 3.38 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr12_-_25796463 | 3.38 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr23_+_23730717 | 3.36 |
|
|
|
| chr4_-_17066886 | 3.36 |
ENSDART00000134595
|
sox5
|
SRY (sex determining region Y)-box 5 |
| KN150405v1_-_65814 | 3.34 |
ENSDART00000160124
|
CABZ01076275.1
|
ENSDARG00000100753 |
| chr17_-_35934175 | 3.32 |
ENSDART00000110040
ENSDART00000137525 |
sox11a
AL929378.1
|
SRY (sex determining region Y)-box 11a ENSDARG00000092907 |
| chr12_+_25509394 | 3.31 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr12_-_1442400 | 3.31 |
|
|
|
| chr16_-_36022327 | 3.28 |
ENSDART00000172252
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr5_-_14148028 | 3.28 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr20_-_25063937 | 3.27 |
ENSDART00000159122
|
epha7
|
eph receptor A7 |
| chr13_-_5440923 | 3.25 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr13_+_7575753 | 3.24 |
|
|
|
| chr1_+_49894978 | 3.24 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr23_+_23559246 | 3.23 |
ENSDART00000172214
|
agrn
|
agrin |
| chr23_+_33362843 | 3.22 |
|
|
|
| chr19_+_41924878 | 3.22 |
ENSDART00000172311
|
si:ch211-57n23.1
|
si:ch211-57n23.1 |
| chr2_+_23139845 | 3.21 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr3_+_36146072 | 3.20 |
ENSDART00000148444
|
zgc:86896
|
zgc:86896 |
| chr4_-_16556116 | 3.17 |
ENSDART00000033188
|
btg1
|
B-cell translocation gene 1, anti-proliferative |
| chr2_-_42384993 | 3.17 |
ENSDART00000141358
|
apom
|
apolipoprotein M |
| chr9_-_34129128 | 3.16 |
|
|
|
| chr7_+_22386456 | 3.16 |
ENSDART00000146081
|
ponzr5
|
plac8 onzin related protein 5 |
| chr16_-_22797736 | 3.13 |
ENSDART00000143836
|
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr23_-_26798851 | 3.07 |
ENSDART00000170576
ENSDART00000166123 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr7_-_54407461 | 3.06 |
ENSDART00000162795
|
ccnd1
|
cyclin D1 |
| chr4_+_22959458 | 3.04 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr14_-_24094142 | 3.04 |
ENSDART00000126894
|
fam13b
|
family with sequence similarity 13, member B |
| chr4_+_14361566 | 3.02 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr21_-_3301751 | 3.01 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr13_-_42630425 | 3.00 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr4_+_14972481 | 2.99 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr12_-_1442655 | 2.98 |
|
|
|
| chr3_-_49242655 | 2.98 |
ENSDART00000142200
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr1_-_35196892 | 2.97 |
ENSDART00000033566
|
smad1
|
SMAD family member 1 |
| chr24_-_23175007 | 2.97 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr6_+_58921655 | 2.96 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr5_-_68270398 | 2.95 |
ENSDART00000161561
|
ENSDARG00000059093
|
ENSDARG00000059093 |
| chr1_-_46047321 | 2.93 |
|
|
|
| chr13_+_25298383 | 2.92 |
|
|
|
| chr12_+_40919018 | 2.92 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr17_-_30846176 | 2.88 |
ENSDART00000062811
|
yy1a
|
YY1 transcription factor a |
| chr8_+_47861033 | 2.87 |
ENSDART00000147159
|
plod1a
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1a |
| chr4_+_14361655 | 2.86 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr6_-_39315608 | 2.85 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr18_+_29424354 | 2.85 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr1_-_29669484 | 2.83 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr20_+_13998066 | 2.83 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr17_+_52736192 | 2.82 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr9_-_14533551 | 2.80 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr1_+_29054033 | 2.80 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr3_+_30790513 | 2.80 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr16_+_21098948 | 2.79 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr20_-_34389652 | 2.78 |
ENSDART00000144705
ENSDART00000020389 |
hmcn1
|
hemicentin 1 |
| chr19_+_5375413 | 2.77 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr18_-_15404998 | 2.74 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr7_+_34417030 | 2.73 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr24_+_7832020 | 2.72 |
ENSDART00000019705
|
bmp6
|
bone morphogenetic protein 6 |
| chr10_-_43924675 | 2.72 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr21_+_7844259 | 2.71 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr9_-_33584190 | 2.71 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr18_-_46260401 | 2.70 |
ENSDART00000153930
|
si:dkey-244a7.1
|
si:dkey-244a7.1 |
| chr3_-_34363698 | 2.70 |
|
|
|
| chr15_-_11257445 | 2.69 |
|
|
|
| chr7_-_56493164 | 2.69 |
|
|
|
| chr11_-_22438900 | 2.69 |
ENSDART00000014062
|
myog
|
myogenin |
| chr21_-_23074223 | 2.68 |
ENSDART00000147896
|
usp28
|
ubiquitin specific peptidase 28 |
| chr13_-_3022033 | 2.67 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr9_-_24183317 | 2.67 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr23_-_18972097 | 2.66 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr16_-_13734068 | 2.65 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr20_-_47521258 | 2.65 |
|
|
|
| chr12_+_18112818 | 2.64 |
ENSDART00000130741
|
fam20cb
|
family with sequence similarity 20, member Cb |
| chr25_+_558191 | 2.64 |
ENSDART00000126863
ENSDART00000166012 |
nell2a
|
neural EGFL like 2a |
| chr24_-_23174888 | 2.63 |
ENSDART00000112256
|
zfhx4
|
zinc finger homeobox 4 |
| chr16_-_13733759 | 2.63 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr17_-_52735250 | 2.63 |
|
|
|
| chr18_-_14723138 | 2.63 |
ENSDART00000010129
|
pdf
|
peptide deformylase (mitochondrial) |
| chr5_-_71340996 | 2.62 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr11_+_37639045 | 2.62 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr20_+_26639029 | 2.62 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr23_-_3466041 | 2.61 |
ENSDART00000002309
|
mafba
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
| chr20_+_29731714 | 2.59 |
ENSDART00000101556
|
asap2b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2b |
| chr22_-_23238714 | 2.58 |
ENSDART00000054807
|
lhx9
|
LIM homeobox 9 |
| chr9_-_1958795 | 2.58 |
ENSDART00000138730
|
hoxd3a
|
homeobox D3a |
| chr7_-_41347831 | 2.58 |
ENSDART00000174304
|
plxdc2
|
plexin domain containing 2 |
| chr7_+_73598802 | 2.57 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr8_-_40287789 | 2.57 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr25_+_31053243 | 2.56 |
ENSDART00000067039
|
rassf8a
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8a |
| chr4_+_7668691 | 2.55 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr16_+_39396358 | 2.55 |
|
|
|
| KN149795v1_-_24393 | 2.54 |
|
|
|
| chr15_-_27778111 | 2.53 |
ENSDART00000134373
|
lhx1a
|
LIM homeobox 1a |
| chr20_+_34423970 | 2.53 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr1_-_55554518 | 2.52 |
ENSDART00000019573
|
zgc:65894
|
zgc:65894 |
| chr7_+_30516734 | 2.52 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr14_+_23980348 | 2.51 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr23_-_18958008 | 2.51 |
ENSDART00000133419
|
CR847953.1
|
ENSDARG00000057403 |
| chr3_-_32464649 | 2.50 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr20_+_34640226 | 2.50 |
ENSDART00000076946
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr21_-_11539798 | 2.49 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr6_-_59510397 | 2.47 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr7_+_39130411 | 2.46 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr6_-_27901354 | 2.46 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr9_-_9864049 | 2.45 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr22_+_2921305 | 2.44 |
ENSDART00000143258
|
cep19
|
centrosomal protein 19 |
| chr2_+_25363717 | 2.44 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr14_+_6122948 | 2.43 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr2_+_3701942 | 2.42 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr2_-_30071872 | 2.41 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr7_+_25794014 | 2.40 |
ENSDART00000174010
ENSDART00000173815 |
CT027989.1
|
ENSDARG00000105638 |
| chr6_-_39053400 | 2.39 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr23_-_19789409 | 2.39 |
ENSDART00000131860
|
rpl10
|
ribosomal protein L10 |
| chr6_-_49527616 | 2.38 |
ENSDART00000128025
|
rps26l
|
ribosomal protein S26, like |
| chr18_-_22297945 | 2.37 |
|
|
|
| chr11_+_24687813 | 2.37 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr7_-_33080261 | 2.36 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr4_+_18974767 | 2.36 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr5_+_32739678 | 2.36 |
ENSDART00000026085
|
ptges
|
prostaglandin E synthase |
| chr1_-_46015367 | 2.36 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr19_+_7798512 | 2.35 |
ENSDART00000104719
ENSDART00000146747 |
tuft1b
|
tuftelin 1b |
| chr13_+_27102308 | 2.34 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr7_-_33678058 | 2.34 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr2_-_30071993 | 2.33 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr6_+_49096966 | 2.32 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr13_+_28364942 | 2.32 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr1_-_10201135 | 2.32 |
ENSDART00000131744
|
dmd
|
dystrophin |
| chr1_-_45709828 | 2.31 |
ENSDART00000150029
|
atp11a
|
ATPase, class VI, type 11A |
| chr1_-_10822278 | 2.30 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr1_-_44888989 | 2.30 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_+_43018443 | 2.30 |
ENSDART00000064888
|
tcta
|
T-cell leukemia translocation altered gene |
| chr16_-_35633520 | 2.30 |
ENSDART00000169868
ENSDART00000166606 |
scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr11_-_23151247 | 2.29 |
|
|
|
| chr3_-_58170499 | 2.29 |
ENSDART00000074272
|
im:6904045
|
im:6904045 |
| chr12_-_25973094 | 2.29 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr11_-_33355651 | 2.29 |
ENSDART00000033980
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr17_-_20124053 | 2.27 |
ENSDART00000133327
|
actn2b
|
actinin, alpha 2b |
| chr9_-_12604530 | 2.26 |
ENSDART00000128931
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr2_+_27739201 | 2.26 |
ENSDART00000145909
|
sepp1b
|
selenoprotein P, plasma, 1b |
| chr5_+_34397578 | 2.26 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr15_+_7179382 | 2.25 |
ENSDART00000101578
|
her8.2
|
hairy-related 8.2 |
| chr25_-_31452381 | 2.25 |
ENSDART00000028338
|
scamp5a
|
secretory carrier membrane protein 5a |
| chr21_+_25651712 | 2.25 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0097377 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 1.7 | 5.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 1.6 | 8.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 1.5 | 5.9 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 1.4 | 4.2 | GO:0097702 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 1.4 | 4.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 1.4 | 4.2 | GO:0045933 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 1.3 | 4.0 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 1.3 | 5.3 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 1.3 | 6.6 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 1.2 | 5.0 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 1.1 | 3.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 1.1 | 4.4 | GO:0061194 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 1.0 | 5.1 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 1.0 | 3.0 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 1.0 | 4.8 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.9 | 2.8 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.9 | 3.7 | GO:1901741 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.9 | 5.4 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.9 | 2.6 | GO:0070166 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.9 | 4.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.7 | 4.5 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.7 | 6.4 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.7 | 4.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.7 | 2.8 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.7 | 2.7 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.7 | 2.0 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.7 | 5.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.6 | 1.9 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.6 | 1.9 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.6 | 15.5 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.6 | 4.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.6 | 2.5 | GO:0036363 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.6 | 11.1 | GO:0032963 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.6 | 9.8 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.6 | 2.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.6 | 1.8 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.6 | 4.2 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.6 | 3.6 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.6 | 2.3 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.5 | 1.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.5 | 2.2 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.5 | 1.6 | GO:0015822 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.5 | 2.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.5 | 2.6 | GO:0002030 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.5 | 1.5 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.5 | 2.0 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.5 | 2.0 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.5 | 5.7 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.5 | 1.4 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.5 | 1.8 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.4 | 2.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.4 | 2.7 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.4 | 0.9 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.4 | 1.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.4 | 7.4 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.4 | 1.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 1.2 | GO:0097037 | heme export(GO:0097037) |
| 0.4 | 3.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.4 | 0.8 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.4 | 1.5 | GO:0002832 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.4 | 2.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.4 | 5.2 | GO:0014028 | notochord formation(GO:0014028) |
| 0.4 | 1.5 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.4 | 1.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 1.4 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.3 | 2.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.3 | 3.8 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.3 | 1.7 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.3 | 1.4 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 2.4 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.3 | 5.7 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 1.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.3 | 1.6 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.3 | 2.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 1.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.3 | 3.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 2.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.3 | 2.8 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.3 | 3.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.3 | 3.3 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.3 | 1.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.3 | 0.9 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.3 | 2.4 | GO:0097028 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.3 | 1.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 3.1 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.3 | 2.5 | GO:0021794 | thalamus development(GO:0021794) |
| 0.3 | 2.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.3 | 1.1 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.3 | 2.5 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.3 | 3.3 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.3 | 1.9 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 1.3 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.3 | 3.0 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 6.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.3 | 0.8 | GO:0072045 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.3 | 3.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 7.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.3 | 2.6 | GO:0030241 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 1.0 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.3 | 1.8 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 16.2 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.3 | 1.5 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.2 | 1.2 | GO:0099640 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.2 | 1.0 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 1.7 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.2 | 0.7 | GO:0009215 | deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.2 | 3.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 2.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 0.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 2.9 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.7 | GO:0030803 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.2 | 3.9 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 2.2 | GO:0006956 | complement activation(GO:0006956) |
| 0.2 | 6.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.2 | 1.9 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.2 | 5.1 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.2 | 1.0 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 0.6 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 1.6 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 8.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.2 | 1.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 2.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.4 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.2 | 2.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 4.5 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 2.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.2 | 0.8 | GO:0046636 | negative regulation of alpha-beta T cell activation(GO:0046636) negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.2 | 1.7 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.2 | 0.7 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.2 | 0.7 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.2 | 0.7 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 0.4 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.2 | 2.6 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.2 | 1.4 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.2 | 3.8 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.2 | 0.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.2 | 1.0 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.2 | 1.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 0.8 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 1.2 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 1.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 12.9 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.2 | 2.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 0.8 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.2 | 1.1 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.2 | 1.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 2.6 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.2 | 0.5 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.2 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 3.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 1.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 2.6 | GO:0001819 | positive regulation of cytokine production(GO:0001819) |
| 0.1 | 1.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 3.6 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 1.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 3.8 | GO:0036303 | lymph vessel morphogenesis(GO:0036303) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.4 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 2.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.9 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 1.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 6.9 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 2.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.3 | GO:0072506 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) phosphate ion homeostasis(GO:0055062) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 5.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.9 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.6 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 4.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.4 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.1 | 1.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.7 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.1 | 4.0 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 2.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 3.0 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.2 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.0 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 1.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 1.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 2.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 0.4 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.1 | 1.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.9 | GO:0060324 | face development(GO:0060324) |
| 0.1 | 1.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.2 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 1.4 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.1 | 0.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.2 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 1.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.1 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 1.0 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.1 | 1.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.1 | 1.6 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.1 | 0.9 | GO:0003146 | heart jogging(GO:0003146) |
| 0.1 | 1.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 1.0 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 9.9 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.1 | 0.6 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.8 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 1.9 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.8 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 1.6 | GO:2000181 | negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.1 | 2.2 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.1 | 1.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 0.4 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.1 | 1.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.1 | 2.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 1.3 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.8 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.7 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.7 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 4.5 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.2 | GO:0072530 | purine-containing compound transmembrane transport(GO:0072530) |
| 0.0 | 1.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 2.6 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 0.5 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0006555 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.8 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.5 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 2.0 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.3 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.8 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 2.0 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.2 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.0 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.9 | GO:2001056 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) positive regulation of cysteine-type endopeptidase activity(GO:2001056) |
| 0.0 | 0.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 1.3 | GO:0097485 | axon guidance(GO:0007411) neuron projection guidance(GO:0097485) |
| 0.0 | 0.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0043666 | regulation of phosphatase activity(GO:0010921) regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 1.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.3 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.1 | 6.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.7 | 2.8 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.7 | 2.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.6 | 35.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.6 | 1.9 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.6 | 2.5 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.6 | 4.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.6 | 7.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.5 | 12.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 3.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.4 | 1.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 7.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 1.0 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 3.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.3 | 4.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.3 | 5.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.3 | 4.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 2.9 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.3 | 1.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 2.6 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 11.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.2 | 1.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.2 | 1.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 2.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.2 | 6.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.2 | 3.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 1.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.2 | 1.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 6.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 9.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 8.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.2 | 1.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 1.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 1.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.2 | 0.8 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.2 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 22.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 6.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.4 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 2.0 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 4.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 1.7 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 63.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 1.8 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 3.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 3.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.4 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.4 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 1.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.8 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 3.3 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 3.7 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 7.8 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 1.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 3.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 5.7 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 5.1 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 2.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 7.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.0 | GO:0060076 | postsynaptic density(GO:0014069) excitatory synapse(GO:0060076) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 9.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.6 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 2.0 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0030684 | preribosome(GO:0030684) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.9 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.6 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 1.2 | 4.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.2 | 4.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 1.1 | 3.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.9 | 2.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.9 | 7.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.9 | 4.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.8 | 6.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.7 | 7.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.7 | 2.1 | GO:0070699 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.7 | 10.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.7 | 5.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.6 | 5.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.6 | 1.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.6 | 1.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 2.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.6 | 1.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.6 | 7.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.5 | 1.6 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.5 | 1.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.5 | 1.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.5 | 9.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.5 | 1.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 3.5 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.5 | 2.0 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.5 | 7.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 2.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.5 | 2.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 1.8 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.4 | 26.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 2.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.4 | 1.3 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.4 | 2.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.4 | 4.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.4 | 2.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 1.1 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.4 | 2.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.4 | 3.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 1.0 | GO:0070548 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.3 | 4.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.3 | 2.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 2.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.3 | 1.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 2.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 1.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.3 | 0.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 2.5 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 3.0 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.3 | 1.6 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 3.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 0.8 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.3 | 4.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.8 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 1.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.2 | 0.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 4.0 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 3.2 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.2 | 1.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.7 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 8.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 1.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 1.0 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 2.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 1.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 2.9 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.2 | 1.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 2.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.4 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.7 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.2 | 2.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 3.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.2 | 0.5 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 13.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.2 | 0.8 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 1.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.2 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 2.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 9.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 11.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 15.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 1.0 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 1.0 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 17.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 108.8 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.1 | 0.3 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 0.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.5 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.3 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.5 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 7.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.3 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 2.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.8 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.4 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 15.9 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.1 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.1 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.9 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 0.9 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.1 | 0.8 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.5 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.3 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 5.0 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 21.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 2.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 2.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 2.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 0.5 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.7 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.7 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 1.8 | GO:0005216 | ion channel activity(GO:0005216) |
| 0.0 | 0.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 17.7 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.6 | 6.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 3.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.6 | 4.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.6 | 1.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.5 | 10.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 1.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 2.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.3 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 4.3 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.3 | 3.0 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.0 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 9.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 16.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 2.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 1.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.2 | 3.9 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.8 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 4.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.9 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 4.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 4.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 0.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 1.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.5 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.0 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.7 | 3.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.6 | 3.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.5 | 2.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.4 | 3.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 3.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.4 | 6.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.4 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 3.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 7.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.4 | 2.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 1.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.3 | 7.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 3.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.3 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.3 | 11.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.3 | 1.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.3 | 3.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 4.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 6.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.2 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.2 | 1.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 1.9 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 6.1 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.2 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 7.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 2.0 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 3.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 4.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 11.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.5 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 0.8 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 0.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.5 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.1 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 4.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 5.2 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.9 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.5 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |