Project
DANIO-CODE
Navigation
Downloads
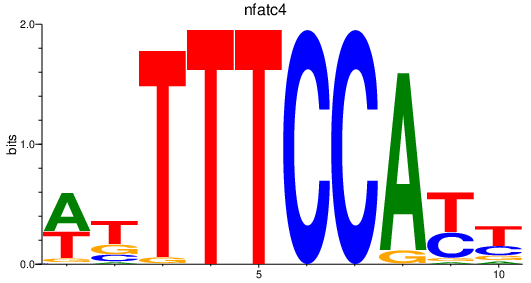
Results for nfatc4
Z-value: 1.63
Transcription factors associated with nfatc4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfatc4
|
ENSDARG00000054162 | nuclear factor of activated T cells 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfatc4 | dr10_dc_chr2_-_37821146_37821201 | -0.77 | 4.2e-04 | Click! |
Activity profile of nfatc4 motif
Sorted Z-values of nfatc4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfatc4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_49880744 | 5.53 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr16_+_23482744 | 4.28 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr2_+_42342148 | 3.96 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr9_-_34129128 | 3.90 |
|
|
|
| chr3_+_26013873 | 3.87 |
ENSDART00000043932
|
atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr22_-_37414940 | 3.63 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr7_-_23751107 | 3.63 |
ENSDART00000149133
|
cideb
|
cell death-inducing DFFA-like effector b |
| chr3_-_19942214 | 3.63 |
ENSDART00000029386
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr6_-_39315024 | 3.62 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr2_-_41669008 | 3.57 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr1_-_57931565 | 3.55 |
ENSDART00000109528
|
adgre5b.3
|
adhesion G protein-coupled receptor E5b, duplicate 3 |
| chr6_+_14823185 | 3.52 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr12_-_30988279 | 3.51 |
ENSDART00000122972
ENSDART00000005562 ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 |
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_41396014 | 3.39 |
ENSDART00000159444
ENSDART00000042990 ENSDART00000144544 |
col1a2
|
collagen, type I, alpha 2 |
| chr12_-_25973094 | 3.30 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr4_+_17428131 | 3.18 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr25_-_14948922 | 3.14 |
ENSDART00000161165
|
pax6a
|
paired box 6a |
| chr25_-_14472107 | 3.08 |
|
|
|
| chr19_-_41482388 | 3.02 |
ENSDART00000111982
|
sgce
|
sarcoglycan, epsilon |
| chr22_-_26333957 | 2.96 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr13_-_46292762 | 2.93 |
ENSDART00000149125
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr6_+_49096966 | 2.81 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr7_-_52283383 | 2.75 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr21_-_5852663 | 2.72 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr6_+_6333766 | 2.71 |
ENSDART00000140827
|
bcl11ab
|
B-cell CLL/lymphoma 11Ab |
| chr2_+_4366169 | 2.67 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr23_+_36032206 | 2.62 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr14_-_32404076 | 2.61 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr1_+_49894978 | 2.60 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr2_+_38178934 | 2.51 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr14_+_33117964 | 2.49 |
ENSDART00000075278
|
atp1b4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr10_-_43711606 | 2.44 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr6_-_49064365 | 2.43 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr14_-_425659 | 2.41 |
|
|
|
| chr1_-_50831155 | 2.40 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr22_+_16511562 | 2.38 |
|
|
|
| chr14_+_23980348 | 2.36 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr1_-_11591843 | 2.36 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr11_-_26428979 | 2.34 |
ENSDART00000111539
|
efcc1
|
EF-hand and coiled-coil domain containing 1 |
| chr25_+_35179025 | 2.34 |
ENSDART00000149768
|
kif21a
|
kinesin family member 21A |
| chr9_+_53736715 | 2.34 |
ENSDART00000003310
|
sox21b
|
SRY (sex determining region Y)-box 21b |
| chr17_-_45383925 | 2.34 |
|
|
|
| KN150640v1_+_5728 | 2.30 |
|
|
|
| chr2_-_41669059 | 2.30 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr16_+_29558320 | 2.29 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr11_-_23954234 | 2.29 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr14_+_6122948 | 2.26 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr4_-_15442828 | 2.26 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr16_-_17439735 | 2.25 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr7_+_30516734 | 2.23 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr2_+_23139845 | 2.16 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr1_-_37475807 | 2.13 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr9_+_36051713 | 2.11 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr2_-_21265803 | 2.10 |
ENSDART00000006870
|
ptgs2a
|
prostaglandin-endoperoxide synthase 2a |
| chr8_+_25170822 | 2.10 |
ENSDART00000047528
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr6_-_50705420 | 2.09 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr16_-_36022327 | 2.08 |
ENSDART00000172252
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr4_+_10065500 | 2.07 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr15_-_39756101 | 2.07 |
|
|
|
| chr21_-_3301751 | 2.06 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr9_-_1958795 | 2.06 |
ENSDART00000138730
|
hoxd3a
|
homeobox D3a |
| chr13_+_1025144 | 2.05 |
ENSDART00000148356
|
perp
|
PERP, TP53 apoptosis effector |
| chr5_+_6054781 | 2.04 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr5_+_31745031 | 2.02 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr1_-_53746151 | 2.02 |
ENSDART00000179565
|
CABZ01043826.1
|
ENSDARG00000108130 |
| chr4_+_12014166 | 2.02 |
ENSDART00000034401
ENSDART00000172691 |
cry1aa
|
cryptochrome circadian clock 1aa |
| chr13_+_28997608 | 2.00 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr12_+_18112818 | 1.98 |
ENSDART00000130741
|
fam20cb
|
family with sequence similarity 20, member Cb |
| chr8_-_40425193 | 1.98 |
ENSDART00000130798
|
myl7
|
myosin, light chain 7, regulatory |
| chr5_+_34397578 | 1.97 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr6_-_39315608 | 1.97 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr23_+_36002332 | 1.91 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr5_-_43334777 | 1.91 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr20_+_23599157 | 1.88 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr11_+_40652239 | 1.88 |
ENSDART00000169817
ENSDART00000162157 ENSDART00000172008 |
pax7a
|
paired box 7a |
| chr13_+_13550656 | 1.86 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr13_+_28997676 | 1.86 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr4_-_23362106 | 1.84 |
ENSDART00000133644
ENSDART00000009768 |
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr6_-_48095609 | 1.83 |
ENSDART00000137458
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr4_-_22590638 | 1.82 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr4_+_7668939 | 1.81 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr20_+_15653121 | 1.79 |
ENSDART00000152702
|
jun
|
jun proto-oncogene |
| chr20_+_31384654 | 1.79 |
ENSDART00000007688
|
slc22a16
|
solute carrier family 22 (organic cation/carnitine transporter), member 16 |
| chr1_-_4757890 | 1.76 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr9_-_53220424 | 1.76 |
|
|
|
| chr4_-_9721600 | 1.74 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr1_-_11591684 | 1.72 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr13_-_46292975 | 1.72 |
ENSDART00000167402
ENSDART00000080914 ENSDART00000080916 |
fgfr2
|
fibroblast growth factor receptor 2 |
| chr23_-_24755654 | 1.71 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr10_-_43924675 | 1.70 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr25_-_14949028 | 1.69 |
ENSDART00000172538
|
pax6a
|
paired box 6a |
| chr5_+_15319430 | 1.68 |
ENSDART00000162003
|
hspb8
|
heat shock protein b8 |
| chr17_+_24830545 | 1.68 |
ENSDART00000062917
|
cx35.4
|
connexin 35.4 |
| chr3_+_23591228 | 1.64 |
ENSDART00000012470
|
hoxb4a
|
homeobox B4a |
| chr16_-_13733759 | 1.63 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr15_-_25582891 | 1.62 |
|
|
|
| chr15_-_1858350 | 1.62 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr18_-_23889256 | 1.62 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_+_18639896 | 1.61 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr3_-_39245184 | 1.61 |
|
|
|
| chr18_-_36291570 | 1.60 |
|
|
|
| chr13_-_28177612 | 1.59 |
ENSDART00000045351
|
lbx1a
|
ladybird homeobox 1a |
| chr9_-_24183317 | 1.59 |
ENSDART00000140346
|
col6a3
|
collagen, type VI, alpha 3 |
| chr12_-_9479063 | 1.59 |
ENSDART00000169727
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| KN150349v1_-_13313 | 1.58 |
|
|
|
| chr16_-_22797736 | 1.58 |
ENSDART00000143836
|
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr3_+_21928483 | 1.57 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr1_+_45503061 | 1.56 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr16_+_21098948 | 1.56 |
ENSDART00000006043
|
hoxa11b
|
homeobox A11b |
| chr11_-_18542803 | 1.55 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr9_-_46272322 | 1.55 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr4_+_2215426 | 1.54 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr8_+_30491476 | 1.52 |
ENSDART00000062073
|
ENSDARG00000042329
|
ENSDARG00000042329 |
| chr18_-_605558 | 1.51 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr10_+_4717800 | 1.51 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr2_-_37974443 | 1.49 |
ENSDART00000034595
|
cbln10
|
cerebellin 10 |
| chr8_+_48488009 | 1.49 |
|
|
|
| chr11_-_37158131 | 1.48 |
ENSDART00000160911
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr11_-_32461160 | 1.48 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr12_+_46944395 | 1.48 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr18_+_23891068 | 1.47 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr3_-_19942332 | 1.47 |
ENSDART00000124326
|
slc4a1a
|
solute carrier family 4 (anion exchanger), member 1a (Diego blood group) |
| chr6_-_49674729 | 1.47 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr6_+_53349584 | 1.46 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr1_-_22144014 | 1.45 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr3_+_28871566 | 1.43 |
|
|
|
| chr7_-_52142689 | 1.42 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr9_+_3416525 | 1.42 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr1_-_40489256 | 1.41 |
ENSDART00000084665
|
dok7
|
docking protein 7 |
| chr10_-_43711513 | 1.41 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr17_+_22082952 | 1.38 |
ENSDART00000047772
|
mal
|
mal, T-cell differentiation protein |
| KN150131v1_+_59098 | 1.37 |
ENSDART00000175590
|
CABZ01064941.1
|
ENSDARG00000106550 |
| chr10_+_3728747 | 1.37 |
|
|
|
| chr16_+_21104310 | 1.36 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr24_+_11194381 | 1.33 |
ENSDART00000143171
|
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr7_-_26036887 | 1.33 |
ENSDART00000146935
|
zgc:77439
|
zgc:77439 |
| chr10_-_31619761 | 1.33 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr7_-_55214437 | 1.32 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr16_+_9141325 | 1.31 |
|
|
|
| chr11_-_34367105 | 1.30 |
ENSDART00000157625
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr12_+_40919018 | 1.30 |
ENSDART00000176047
|
BX072562.1
|
ENSDARG00000108020 |
| chr10_-_38372282 | 1.30 |
ENSDART00000149580
|
nrip1b
|
nuclear receptor interacting protein 1b |
| chr25_+_27050388 | 1.29 |
ENSDART00000112299
|
gpr37a
|
G protein-coupled receptor 37a |
| chr15_-_6417147 | 1.28 |
|
|
|
| chr4_-_23362476 | 1.27 |
ENSDART00000133644
ENSDART00000009768 |
magi2a
|
membrane associated guanylate kinase, WW and PDZ domain containing 2a |
| chr22_+_20436287 | 1.26 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr5_+_37113743 | 1.26 |
|
|
|
| chr17_+_48999061 | 1.26 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr24_+_16005004 | 1.26 |
ENSDART00000163086
ENSDART00000152087 |
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr7_-_40713381 | 1.25 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr23_-_7892862 | 1.25 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr3_-_30303173 | 1.24 |
ENSDART00000150958
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr9_-_35135414 | 1.24 |
ENSDART00000140563
|
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr18_+_15191737 | 1.24 |
|
|
|
| chr14_+_20862466 | 1.24 |
ENSDART00000059796
|
ENSDARG00000019213
|
ENSDARG00000019213 |
| chr8_+_22910147 | 1.23 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr3_+_47735048 | 1.22 |
ENSDART00000083024
|
gpr146
|
G protein-coupled receptor 146 |
| chr14_-_49951676 | 1.22 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr15_-_20997836 | 1.22 |
ENSDART00000152648
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr4_+_18854068 | 1.21 |
ENSDART00000066977
|
bik
|
BCL2-interacting killer (apoptosis-inducing) |
| chr1_-_46398498 | 1.20 |
ENSDART00000000087
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr2_-_48341913 | 1.20 |
ENSDART00000139944
|
si:dkeyp-12a9.5
|
si:dkeyp-12a9.5 |
| chr2_-_30198789 | 1.19 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr13_-_18704185 | 1.19 |
ENSDART00000146795
|
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr4_+_10618211 | 1.19 |
ENSDART00000067251
|
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr3_-_28078162 | 1.19 |
ENSDART00000165936
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr5_-_40310638 | 1.18 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr8_+_17042976 | 1.17 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr7_+_73598802 | 1.16 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr16_-_13734068 | 1.16 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr9_+_3416651 | 1.15 |
ENSDART00000019910
|
dlx1a
|
distal-less homeobox 1a |
| chr1_+_4557485 | 1.15 |
ENSDART00000052423
|
spry2
|
sprouty RTK signaling antagonist 2 |
| chr20_+_34640226 | 1.14 |
ENSDART00000076946
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr5_-_40310702 | 1.12 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr9_-_24219908 | 1.10 |
ENSDART00000144163
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr21_+_142265 | 1.10 |
|
|
|
| chr2_-_32575527 | 1.10 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr13_-_46292862 | 1.10 |
ENSDART00000150061
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr4_+_19545842 | 1.10 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr14_-_16791447 | 1.10 |
ENSDART00000158002
|
CR812832.1
|
ENSDARG00000103278 |
| chr8_-_34077387 | 1.09 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr22_+_13862110 | 1.09 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr18_+_23891152 | 1.09 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr13_-_26907910 | 1.09 |
ENSDART00000146712
|
ccdc85a
|
coiled-coil domain containing 85A |
| chr18_+_26737365 | 1.09 |
ENSDART00000141672
|
alpk3a
|
alpha-kinase 3a |
| chr13_-_31339870 | 1.08 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr2_-_39052185 | 1.08 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr3_+_34064081 | 1.08 |
|
|
|
| chr10_-_30016761 | 1.07 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr11_+_13572592 | 1.07 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr11_+_24052512 | 1.07 |
ENSDART00000029695
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr10_-_35293024 | 1.06 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr20_-_10133201 | 1.06 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr15_-_34987450 | 1.05 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr11_-_30792331 | 1.05 |
|
|
|
| chr25_-_1980943 | 1.05 |
ENSDART00000104915
|
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr14_-_46646747 | 1.02 |
|
|
|
| chr21_-_17566461 | 1.00 |
ENSDART00000044078
ENSDART00000168495 ENSDART00000020048 |
gsna
|
gelsolin a |
| chr6_-_8001756 | 1.00 |
ENSDART00000151483
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr18_-_23889025 | 0.99 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) positive regulation of muscle contraction(GO:0045933) |
| 1.3 | 3.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 1.1 | 3.2 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.9 | 3.5 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.9 | 2.6 | GO:0010712 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.9 | 0.9 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.9 | 2.6 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.8 | 2.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.8 | 4.8 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.8 | 2.3 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.7 | 3.6 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.7 | 5.8 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.7 | 2.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.7 | 3.4 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.7 | 2.0 | GO:0070166 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.5 | 1.5 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.5 | 2.5 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.5 | 2.0 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.5 | 1.9 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.4 | 1.3 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.4 | 2.1 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.4 | 5.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.4 | 2.0 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.4 | 2.8 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.4 | 1.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 1.9 | GO:0006956 | complement activation(GO:0006956) |
| 0.4 | 1.5 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.4 | 6.3 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.4 | 1.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.3 | 1.3 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.3 | 1.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 5.9 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.3 | 1.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.6 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.3 | 3.7 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.2 | 6.4 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.2 | 0.6 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.2 | 1.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.2 | 4.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 2.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 1.1 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.2 | 0.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 1.3 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 1.4 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.2 | 1.2 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.1 | 1.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 1.6 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 1.8 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.4 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.4 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 2.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.0 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.3 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 1.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 3.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.1 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 1.6 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 1.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.8 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.1 | 2.7 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.1 | 0.5 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.8 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 1.5 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 2.1 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 0.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.8 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 4.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 1.2 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 4.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 7.8 | GO:0007187 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.1 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 1.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 2.3 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.1 | 4.7 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.7 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 1.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 1.6 | GO:0048675 | axon extension(GO:0048675) |
| 0.0 | 0.9 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.9 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.9 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 2.9 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 2.1 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 1.1 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.2 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 2.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 1.2 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 0.2 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 6.3 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.2 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 1.0 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.7 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 2.8 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.9 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.9 | GO:0007422 | peripheral nervous system development(GO:0007422) |
| 0.0 | 2.2 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0071594 | B cell differentiation(GO:0030183) T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 2.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0050769 | positive regulation of neurogenesis(GO:0050769) |
| 0.0 | 0.6 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.1 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.9 | 3.5 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.5 | 3.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 2.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 2.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 2.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 2.5 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.3 | 4.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 0.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 1.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 3.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 4.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.5 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 2.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 7.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 5.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.0 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.4 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 5.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 2.5 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 0.4 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 3.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 14.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 3.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 8.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 16.7 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.0 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 7.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.7 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.6 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0072379 | ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.7 | 2.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.5 | 5.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.4 | 2.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 5.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 3.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.4 | 2.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 1.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.4 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 5.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.3 | 3.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 0.8 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.3 | 1.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.3 | 0.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.3 | 1.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 5.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 0.8 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.3 | 1.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.3 | 1.8 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.9 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.2 | 2.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 3.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.2 | 3.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 1.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 1.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 1.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 6.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.7 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0030151 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.1 | 1.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 2.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.5 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 1.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.9 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 2.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 3.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 6.8 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.1 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 5.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 38.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 2.5 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 4.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.4 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 8.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.5 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.2 | 5.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 3.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.2 | 2.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 1.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 5.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 1.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 3.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 3.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.0 | 5.8 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.6 | 2.5 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.6 | 3.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.5 | 3.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.5 | 1.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.5 | 2.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 2.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.2 | 2.0 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 1.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.1 | 2.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.2 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.1 | REACTOME GPCR LIGAND BINDING | Genes involved in GPCR ligand binding |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |