Project
DANIO-CODE
Navigation
Downloads
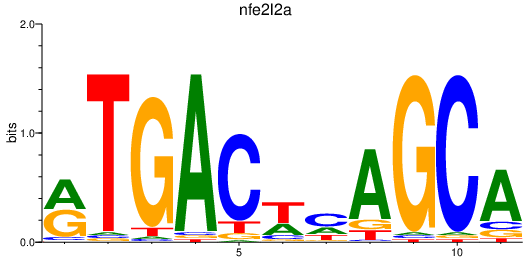
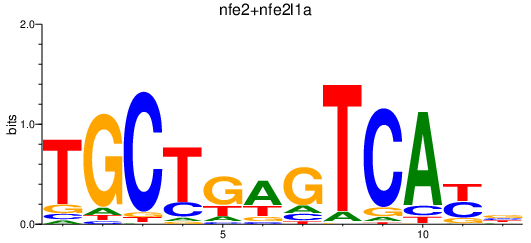
Results for nfe2l2a_nfe2+nfe2l1a
Z-value: 0.48
Transcription factors associated with nfe2l2a_nfe2+nfe2l1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfe2l2a
|
ENSDARG00000042824 | nuclear factor, erythroid 2-like 2a |
|
nfe2
|
ENSDARG00000020606 | nuclear factor, erythroid 2 |
|
nfe2l1a
|
ENSDARG00000030616 | nuclear factor, erythroid 2-like 1a |
Activity profile of nfe2l2a_nfe2+nfe2l1a motif
Sorted Z-values of nfe2l2a_nfe2+nfe2l1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfe2l2a_nfe2+nfe2l1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_66792947 | 0.89 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr5_-_30015572 | 0.68 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr1_-_21961282 | 0.54 |
ENSDART00000171830
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr7_+_61693871 | 0.49 |
|
|
|
| chr20_-_48772949 | 0.47 |
ENSDART00000170894
|
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr24_+_24777962 | 0.46 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr20_-_48773402 | 0.45 |
ENSDART00000161769
|
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr3_-_25886553 | 0.44 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr14_-_25688444 | 0.42 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr5_-_53777106 | 0.39 |
ENSDART00000164067
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr19_-_7531709 | 0.37 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr7_+_17740739 | 0.37 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr22_-_31110564 | 0.36 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr1_+_13386937 | 0.36 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr2_+_25622497 | 0.35 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr23_+_3788435 | 0.35 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr2_+_10023967 | 0.34 |
ENSDART00000148227
|
anxa13l
|
annexin A13, like |
| chr21_+_9483423 | 0.33 |
ENSDART00000162834
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr10_+_38831967 | 0.32 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr5_-_39696678 | 0.32 |
ENSDART00000097526
|
hspb3
|
heat shock protein, alpha-crystallin-related, b3 |
| chr7_+_17740690 | 0.30 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr20_-_14784875 | 0.29 |
ENSDART00000063857
|
scrn2
|
secernin 2 |
| chr20_-_39200252 | 0.28 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr2_-_7693224 | 0.28 |
ENSDART00000167019
|
wu:fc46h12
|
wu:fc46h12 |
| chr7_+_17740815 | 0.28 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr17_-_30884905 | 0.27 |
ENSDART00000131633
ENSDART00000146824 |
evla
|
Enah/Vasp-like a |
| chr11_+_7148830 | 0.27 |
ENSDART00000035560
|
tmem38a
|
transmembrane protein 38A |
| chr10_-_28474085 | 0.27 |
|
|
|
| chr3_-_54738298 | 0.26 |
ENSDART00000145766
|
rhbdf1a
|
rhomboid 5 homolog 1a (Drosophila) |
| chr19_-_30816780 | 0.26 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr3_-_21149752 | 0.26 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr7_+_620117 | 0.25 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr14_+_34440799 | 0.25 |
ENSDART00000173371
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr17_+_8831186 | 0.24 |
ENSDART00000109573
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr21_+_20347283 | 0.24 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr24_-_16772446 | 0.24 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr14_-_38532216 | 0.24 |
ENSDART00000127479
|
gsr
|
glutathione reductase |
| chr19_-_6466494 | 0.23 |
ENSDART00000104950
|
atp1a3a
|
ATPase, Na+/K+ transporting, alpha 3a polypeptide |
| chr20_-_35567985 | 0.22 |
ENSDART00000016090
|
pla2g7
|
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr11_-_29689360 | 0.22 |
ENSDART00000145251
|
pir
|
pirin |
| chr21_-_5228529 | 0.22 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr1_+_50668743 | 0.21 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr7_-_15164722 | 0.21 |
ENSDART00000172147
|
ENSDARG00000036809
|
ENSDARG00000036809 |
| chr20_-_40470331 | 0.21 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr3_+_25992836 | 0.20 |
ENSDART00000010477
|
hsp70.3
|
heat shock cognate 70-kd protein, tandem duplicate 3 |
| chr21_-_35806638 | 0.20 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr18_+_14369543 | 0.20 |
|
|
|
| chr10_+_38831925 | 0.20 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr25_-_16685648 | 0.20 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr4_-_16417703 | 0.19 |
ENSDART00000013085
|
dcn
|
decorin |
| chr2_-_36058327 | 0.19 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr5_-_44243476 | 0.18 |
ENSDART00000161408
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr7_-_71566426 | 0.18 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr10_+_26786051 | 0.18 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr10_+_28420109 | 0.18 |
ENSDART00000142016
|
ptrh2
|
peptidyl-tRNA hydrolase 2 |
| chr7_-_32562435 | 0.17 |
ENSDART00000099872
ENSDART00000099871 ENSDART00000147554 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr9_+_28877812 | 0.17 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr7_-_24337079 | 0.17 |
|
|
|
| chr19_-_18199573 | 0.16 |
ENSDART00000166313
|
thrb
|
thyroid hormone receptor beta |
| chr9_+_41301317 | 0.16 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr13_-_2296317 | 0.16 |
ENSDART00000017148
|
gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr14_+_4044357 | 0.16 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr24_-_16772614 | 0.16 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr10_-_29386085 | 0.16 |
ENSDART00000022605
|
tmem126a
|
transmembrane protein 126A |
| chr14_+_24543732 | 0.16 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr17_+_44992947 | 0.16 |
ENSDART00000156971
|
gstz1
|
glutathione S-transferase zeta 1 |
| chr5_-_53776769 | 0.16 |
ENSDART00000164595
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr23_+_44729796 | 0.15 |
ENSDART00000139088
ENSDART00000139547 |
trappc1
|
trafficking protein particle complex 1 |
| chr7_-_23623141 | 0.15 |
|
|
|
| chr17_-_32826829 | 0.15 |
ENSDART00000077459
|
smyd2a
|
SET and MYND domain containing 2a |
| chr14_+_34440699 | 0.14 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr17_-_36948926 | 0.14 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr15_-_19162052 | 0.14 |
|
|
|
| chr3_+_56882467 | 0.14 |
|
|
|
| chr10_-_28474052 | 0.14 |
|
|
|
| chr7_+_10346939 | 0.14 |
|
|
|
| chr5_-_31328823 | 0.14 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr2_-_6203659 | 0.14 |
ENSDART00000164269
|
prdx1
|
peroxiredoxin 1 |
| chr7_-_39281312 | 0.14 |
ENSDART00000170900
|
slc22a18
|
solute carrier family 22, member 18 |
| chr18_+_1583720 | 0.14 |
|
|
|
| chr16_+_25344184 | 0.14 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| KN150349v1_-_1437 | 0.14 |
|
|
|
| chr20_-_1293519 | 0.13 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr2_-_1677190 | 0.13 |
ENSDART00000024135
|
tubb2
|
tubulin, beta 2A class IIa |
| chr3_-_54884533 | 0.13 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr14_+_29133320 | 0.13 |
ENSDART00000177215
|
ENSDARG00000037122
|
ENSDARG00000037122 |
| chr19_+_10477101 | 0.13 |
ENSDART00000151735
|
necap1
|
NECAP endocytosis associated 1 |
| chr21_-_23074223 | 0.13 |
ENSDART00000147896
|
usp28
|
ubiquitin specific peptidase 28 |
| chr14_+_28158771 | 0.13 |
ENSDART00000019003
|
psmd10
|
proteasome 26S subunit, non-ATPase 10 |
| chr19_+_31989527 | 0.13 |
ENSDART00000088401
|
acot13
|
acyl-CoA thioesterase 13 |
| chr14_-_49600662 | 0.13 |
ENSDART00000161147
|
si:ch211-154c21.1
|
si:ch211-154c21.1 |
| chr8_+_15231620 | 0.13 |
ENSDART00000020386
|
gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr17_+_44992653 | 0.12 |
ENSDART00000157338
|
gstz1
|
glutathione S-transferase zeta 1 |
| chr5_-_53777415 | 0.12 |
ENSDART00000169270
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr19_-_33013748 | 0.12 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr17_-_50899135 | 0.12 |
|
|
|
| chr1_-_32647357 | 0.12 |
ENSDART00000136383
|
cd99
|
CD99 molecule |
| chr12_+_17032963 | 0.12 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr13_-_36164801 | 0.12 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr5_+_65267043 | 0.12 |
ENSDART00000161578
|
mymk
|
myomaker, myoblast fusion factor |
| chr11_+_29543388 | 0.12 |
ENSDART00000067822
|
dynlt3
|
dynein, light chain, Tctex-type 3 |
| chr3_+_5125139 | 0.12 |
ENSDART00000161755
|
CU681842.1
|
ENSDARG00000102795 |
| chr5_-_40894631 | 0.12 |
ENSDART00000121840
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr24_+_9073790 | 0.12 |
|
|
|
| chr7_-_73592082 | 0.12 |
ENSDART00000160951
|
hist1h4l
|
histone 1, H4, like |
| chr11_-_3266080 | 0.11 |
ENSDART00000154314
|
prph
|
peripherin |
| chr7_+_41033728 | 0.11 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr3_+_45345291 | 0.11 |
|
|
|
| chr24_-_28150352 | 0.11 |
ENSDART00000145290
|
bcl2a
|
B-cell CLL/lymphoma 2a |
| chr3_-_30833046 | 0.11 |
|
|
|
| chr3_-_21149711 | 0.11 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr12_+_47029281 | 0.11 |
|
|
|
| chr25_+_5695630 | 0.11 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr1_-_25225339 | 0.11 |
ENSDART00000109714
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr11_+_44922796 | 0.11 |
ENSDART00000172238
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr13_-_36164510 | 0.11 |
ENSDART00000169768
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr21_+_5887486 | 0.11 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr17_+_34168095 | 0.11 |
|
|
|
| chr6_+_36862078 | 0.10 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr20_-_17006936 | 0.10 |
ENSDART00000012859
|
psma6b
|
proteasome subunit alpha 6b |
| chr19_+_2834491 | 0.10 |
ENSDART00000112414
|
rapgef5a
|
Rap guanine nucleotide exchange factor (GEF) 5a |
| chr19_-_7531649 | 0.10 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr20_-_31524684 | 0.10 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| KN149704v1_+_15971 | 0.10 |
|
|
|
| chr23_-_15013317 | 0.10 |
ENSDART00000054909
|
gss
|
glutathione synthetase |
| chr5_-_62816208 | 0.10 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr4_-_12863235 | 0.10 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr21_-_41001910 | 0.09 |
ENSDART00000142277
|
plac8l1
|
PLAC8-like 1 |
| chr23_+_36019758 | 0.09 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr19_+_41630199 | 0.09 |
|
|
|
| chr14_+_15788292 | 0.09 |
ENSDART00000176486
|
immt
|
inner membrane protein, mitochondrial (mitofilin) |
| chr16_+_45964212 | 0.09 |
ENSDART00000128840
ENSDART00000041811 |
otud7b
|
OTU deubiquitinase 7B |
| chr19_-_19453029 | 0.09 |
ENSDART00000163359
ENSDART00000167951 |
dync1li1
|
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr7_-_23474701 | 0.09 |
ENSDART00000048050
|
ITGB1BP2
|
integrin subunit beta 1 binding protein 2 |
| chr24_+_24778026 | 0.09 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr7_-_2308580 | 0.09 |
ENSDART00000098148
ENSDART00000173624 |
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr11_-_25181234 | 0.09 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr15_+_15920149 | 0.09 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr7_-_23623275 | 0.09 |
ENSDART00000123203
|
vbp1
|
von Hippel-Lindau binding protein 1 |
| chr3_+_61903372 | 0.08 |
ENSDART00000108945
|
GID4
|
GID complex subunit 4 homolog |
| chr14_-_2819498 | 0.08 |
ENSDART00000090196
|
slc35a4
|
solute carrier family 35, member A4 |
| chr17_-_49931552 | 0.08 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr12_+_17032997 | 0.08 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr17_+_27157965 | 0.08 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr5_+_19703933 | 0.08 |
|
|
|
| chr23_+_24999817 | 0.08 |
ENSDART00000137486
|
klhl21
|
kelch-like family member 21 |
| chr24_-_37022879 | 0.08 |
ENSDART00000079233
|
dnaaf3l
|
dynein, axonemal, assembly factor 3 like |
| chr12_+_48306867 | 0.08 |
|
|
|
| KN150307v1_+_6450 | 0.08 |
ENSDART00000159959
|
CABZ01038161.1
|
ENSDARG00000098784 |
| chr14_-_30536363 | 0.07 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr12_+_1445117 | 0.07 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr9_-_29769100 | 0.07 |
ENSDART00000140876
|
cenpj
|
centromere protein J |
| chr17_+_32412739 | 0.07 |
ENSDART00000156197
|
CR627496.1
|
ENSDARG00000097538 |
| chr16_-_32628744 | 0.07 |
ENSDART00000137936
|
fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr24_+_36022833 | 0.07 |
ENSDART00000088480
|
abcd4
|
ATP-binding cassette, sub-family D (ALD), member 4 |
| chr19_+_33506239 | 0.07 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr10_-_25737220 | 0.07 |
ENSDART00000135058
|
sod1
|
superoxide dismutase 1, soluble |
| chr7_-_40359613 | 0.07 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr8_+_48954232 | 0.07 |
ENSDART00000132035
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr6_+_60130210 | 0.07 |
ENSDART00000148557
|
aurka
|
aurora kinase A |
| chr1_-_30925005 | 0.06 |
ENSDART00000142296
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr23_+_5632124 | 0.06 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr8_+_26352980 | 0.06 |
ENSDART00000053460
ENSDART00000086763 |
nat6
hyal3
|
N-acetyltransferase 6 hyaluronoglucosaminidase 3 |
| chr9_+_21581061 | 0.06 |
|
|
|
| chr17_-_14956738 | 0.06 |
|
|
|
| chr5_-_39696923 | 0.06 |
ENSDART00000097526
|
hspb3
|
heat shock protein, alpha-crystallin-related, b3 |
| chr4_+_9630077 | 0.06 |
ENSDART00000142286
ENSDART00000138173 |
BX901962.1
|
ENSDARG00000093445 |
| chr10_+_9764591 | 0.06 |
ENSDART00000091780
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr25_+_10388228 | 0.06 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr25_+_5695588 | 0.06 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr23_+_9625994 | 0.06 |
ENSDART00000081433
|
adrm1
|
adhesion regulating molecule 1 |
| chr4_-_12862869 | 0.06 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr16_-_54638972 | 0.06 |
|
|
|
| chr22_-_4622401 | 0.06 |
ENSDART00000106166
|
rx1
|
retinal homeobox gene 1 |
| chr7_-_23623461 | 0.06 |
ENSDART00000123203
|
vbp1
|
von Hippel-Lindau binding protein 1 |
| chr20_+_38821682 | 0.06 |
ENSDART00000015095
|
uts1
|
urotensin 1 |
| chr17_+_23709676 | 0.06 |
ENSDART00000179026
|
zgc:91976
|
zgc:91976 |
| chr7_-_47990610 | 0.06 |
ENSDART00000147968
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr2_-_56511961 | 0.06 |
ENSDART00000168160
|
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr7_+_38479571 | 0.06 |
ENSDART00000170486
|
f2
|
coagulation factor II (thrombin) |
| chr6_+_41448993 | 0.06 |
ENSDART00000029553
|
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr8_+_54171992 | 0.06 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr8_+_21101133 | 0.06 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr19_-_33013672 | 0.06 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr5_+_18797669 | 0.05 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr9_+_28878160 | 0.05 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr4_-_12863112 | 0.05 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr14_-_23387483 | 0.05 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr2_+_393439 | 0.05 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr8_+_15231580 | 0.05 |
ENSDART00000020386
|
gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr2_+_58448918 | 0.05 |
ENSDART00000098173
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr11_-_23460219 | 0.05 |
|
|
|
| chr20_+_27104580 | 0.05 |
ENSDART00000152956
|
si:dkey-177p2.18
|
si:dkey-177p2.18 |
| chr2_-_11720585 | 0.05 |
ENSDART00000019392
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr14_+_25927045 | 0.05 |
ENSDART00000168673
ENSDART00000128971 |
gm2a
|
GM2 ganglioside activator |
| chr19_+_27243907 | 0.05 |
ENSDART00000126006
|
FP085398.1
|
ENSDARG00000087611 |
| chr3_-_32699672 | 0.05 |
|
|
|
| chr1_-_44227583 | 0.05 |
ENSDART00000134635
|
si:dkey-9i23.15
|
si:dkey-9i23.15 |
| chr1_-_39509011 | 0.05 |
ENSDART00000146680
|
ENSDARG00000078251
|
ENSDARG00000078251 |
| chr7_-_50642150 | 0.05 |
ENSDART00000022918
|
ankrd46b
|
ankyrin repeat domain 46b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.5 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) membrane depolarization(GO:0051899) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.4 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.3 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.1 | 0.2 | GO:0042671 | thyroid hormone mediated signaling pathway(GO:0002154) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.3 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.0 | 0.3 | GO:0051224 | negative regulation of protein transport(GO:0051224) negative regulation of establishment of protein localization(GO:1904950) |
| 0.0 | 0.3 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.9 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) muscle hypertrophy(GO:0014896) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0051150 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |