Project
DANIO-CODE
Navigation
Downloads
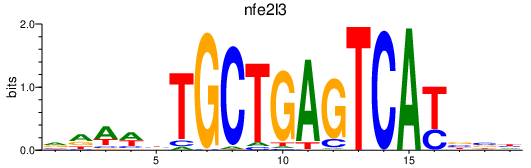
Results for nfe2l3
Z-value: 0.99
Transcription factors associated with nfe2l3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfe2l3
|
ENSDARG00000019810 | nuclear factor, erythroid 2-like 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfe2l3 | dr10_dc_chr19_-_18689571_18689692 | 0.81 | 1.4e-04 | Click! |
Activity profile of nfe2l3 motif
Sorted Z-values of nfe2l3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfe2l3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_39661423 | 2.56 |
ENSDART00000110380
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr14_+_26137608 | 2.43 |
ENSDART00000175459
|
gpr137
|
G protein-coupled receptor 137 |
| chr20_-_17006936 | 2.15 |
ENSDART00000012859
|
psma6b
|
proteasome subunit alpha 6b |
| chr22_-_6977038 | 2.05 |
ENSDART00000133143
ENSDART00000146813 |
gpd1b
|
glycerol-3-phosphate dehydrogenase 1b |
| chr1_-_51862897 | 1.96 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr13_-_22877318 | 1.85 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr3_-_25886553 | 1.74 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr2_+_4366169 | 1.51 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr10_+_29378834 | 1.39 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr12_+_30591157 | 1.34 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr19_-_7235125 | 1.26 |
|
|
|
| chr7_+_56807833 | 1.26 |
ENSDART00000055956
|
enosf1
|
enolase superfamily member 1 |
| chr15_-_44260141 | 1.24 |
ENSDART00000121597
|
CU929391.2
|
ENSDARG00000090304 |
| chr25_-_36876940 | 1.19 |
|
|
|
| chr23_-_44729542 | 1.15 |
ENSDART00000024082
|
psmb6
|
proteasome subunit beta 6 |
| chr10_+_31336284 | 1.13 |
ENSDART00000140988
|
tmem218
|
transmembrane protein 218 |
| chr13_-_22901327 | 1.12 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr14_-_6901415 | 1.10 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr12_-_1931281 | 1.10 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr7_-_6273677 | 1.09 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr14_+_33727750 | 1.04 |
ENSDART00000135556
|
lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr19_+_43524098 | 1.03 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr6_+_9185750 | 1.02 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr21_-_23074223 | 0.97 |
ENSDART00000147896
|
usp28
|
ubiquitin specific peptidase 28 |
| chr6_-_39311711 | 0.96 |
|
|
|
| chr21_+_8249235 | 0.94 |
ENSDART00000129749
|
psmb7
|
proteasome subunit beta 7 |
| chr14_-_25688444 | 0.94 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr18_+_1347470 | 0.93 |
|
|
|
| chr5_-_66792947 | 0.90 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr19_-_6466494 | 0.90 |
ENSDART00000104950
|
atp1a3a
|
ATPase, Na+/K+ transporting, alpha 3a polypeptide |
| chr9_+_41301317 | 0.88 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr20_-_48773402 | 0.88 |
ENSDART00000161769
|
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr19_-_41819752 | 0.86 |
ENSDART00000167772
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr5_-_30015572 | 0.86 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr8_+_39525254 | 0.84 |
|
|
|
| chr8_+_21974863 | 0.81 |
|
|
|
| chr11_-_2107054 | 0.78 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr1_-_51863187 | 0.78 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr2_-_25306278 | 0.75 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr15_-_28744697 | 0.74 |
ENSDART00000060255
|
blmh
|
bleomycin hydrolase |
| chr14_+_34440699 | 0.72 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr20_-_39200252 | 0.72 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr2_-_26987186 | 0.69 |
ENSDART00000132854
|
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr6_+_55413069 | 0.67 |
|
|
|
| chr1_+_52365033 | 0.65 |
ENSDART00000133411
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr16_+_13993746 | 0.65 |
ENSDART00000101304
|
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr15_-_1625846 | 0.65 |
ENSDART00000081875
|
nnr
|
nanor |
| chr6_-_20625494 | 0.65 |
|
|
|
| chr22_+_16509286 | 0.63 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr19_+_25288143 | 0.62 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr6_-_39312306 | 0.59 |
|
|
|
| chr13_+_46487890 | 0.58 |
|
|
|
| chr2_+_4366348 | 0.56 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr14_-_17282615 | 0.55 |
ENSDART00000006716
ENSDART00000136242 |
selt2
|
selenoprotein T, 2 |
| chr14_-_3951077 | 0.54 |
|
|
|
| chr21_-_45308592 | 0.53 |
ENSDART00000008454
|
skp1
|
S-phase kinase-associated protein 1 |
| chr3_-_54884533 | 0.50 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr23_+_5632124 | 0.49 |
ENSDART00000059307
|
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr4_+_5308883 | 0.49 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr17_-_12812747 | 0.48 |
ENSDART00000022874
ENSDART00000046025 |
psma6a
|
proteasome subunit alpha 6a |
| chr3_+_25933164 | 0.48 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr6_-_32362232 | 0.46 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr21_-_25719352 | 0.46 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr14_+_24543732 | 0.45 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr7_+_10346939 | 0.44 |
|
|
|
| chr6_+_59749159 | 0.44 |
|
|
|
| chr1_-_39509011 | 0.43 |
ENSDART00000146680
|
ENSDARG00000078251
|
ENSDARG00000078251 |
| chr5_-_44243476 | 0.42 |
ENSDART00000161408
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr2_-_31817355 | 0.42 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr14_+_34440799 | 0.37 |
ENSDART00000173371
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr14_-_38532216 | 0.36 |
ENSDART00000127479
|
gsr
|
glutathione reductase |
| chr25_-_34677352 | 0.36 |
ENSDART00000115210
|
ano9a
|
anoctamin 9a |
| chr23_+_9625994 | 0.32 |
ENSDART00000081433
|
adrm1
|
adhesion regulating molecule 1 |
| chr7_+_890752 | 0.31 |
ENSDART00000111531
|
epdl1
|
ependymin-like 1 |
| chr1_-_32647357 | 0.31 |
ENSDART00000136383
|
cd99
|
CD99 molecule |
| chr5_+_42312784 | 0.31 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr25_-_34540270 | 0.30 |
ENSDART00000156508
|
zgc:114046
|
zgc:114046 |
| chr7_+_620117 | 0.30 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr23_+_44729796 | 0.30 |
ENSDART00000139088
ENSDART00000139547 |
trappc1
|
trafficking protein particle complex 1 |
| chr25_-_1980943 | 0.29 |
ENSDART00000104915
|
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr4_+_1908127 | 0.29 |
ENSDART00000032460
|
med21
|
mediator complex subunit 21 |
| chr2_+_4366568 | 0.29 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr24_-_3637123 | 0.29 |
|
|
|
| chr9_+_6959065 | 0.28 |
ENSDART00000114323
|
mfsd9
|
major facilitator superfamily domain containing 9 |
| chr11_+_30035395 | 0.27 |
ENSDART00000122756
|
si:dkey-163f14.6
|
si:dkey-163f14.6 |
| chr3_+_15834458 | 0.26 |
|
|
|
| chr9_-_33518171 | 0.26 |
ENSDART00000160534
ENSDART00000132973 |
rpl8
|
ribosomal protein L8 |
| chr7_+_17740815 | 0.26 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr10_+_26786051 | 0.25 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr14_-_6901783 | 0.24 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr3_-_25886604 | 0.23 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr3_-_21149752 | 0.23 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr15_+_44260315 | 0.23 |
ENSDART00000128885
|
cwf19l2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr19_+_43537350 | 0.21 |
|
|
|
| chr7_-_18791134 | 0.20 |
ENSDART00000158796
|
fcer1g
|
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
| chr2_+_4366515 | 0.20 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr16_+_24806508 | 0.20 |
ENSDART00000109459
ENSDART00000155754 |
smg9
|
smg9 nonsense mediated mRNA decay factor |
| chr2_+_10023967 | 0.19 |
ENSDART00000148227
|
anxa13l
|
annexin A13, like |
| chr5_-_23179648 | 0.19 |
|
|
|
| chr18_+_17548213 | 0.19 |
ENSDART00000144960
|
nup93
|
nucleoporin 93 |
| chr18_-_20159345 | 0.19 |
|
|
|
| chr1_+_13386937 | 0.17 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr13_+_25266451 | 0.17 |
ENSDART00000041257
|
gsto2
|
glutathione S-transferase omega 2 |
| chr13_-_41420815 | 0.17 |
ENSDART00000163331
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr8_+_17975195 | 0.16 |
ENSDART00000039887
ENSDART00000121984 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr3_-_21149711 | 0.16 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr22_-_35087752 | 0.15 |
ENSDART00000162211
ENSDART00000158158 |
zgc:63733
|
zgc:63733 |
| chr21_+_27266000 | 0.14 |
ENSDART00000012855
|
sart1
|
squamous cell carcinoma antigen recognised by T cells |
| chr2_+_25622497 | 0.14 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr17_-_11285631 | 0.13 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr9_-_23442944 | 0.10 |
ENSDART00000143657
ENSDART00000169911 ENSDART00000167779 |
acmsd
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr7_-_2308580 | 0.09 |
ENSDART00000098148
ENSDART00000173624 |
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr8_+_39478197 | 0.07 |
|
|
|
| chr14_-_6680528 | 0.07 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr7_-_24337079 | 0.07 |
|
|
|
| chr12_+_30591482 | 0.04 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr5_+_44205128 | 0.04 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr6_-_39312114 | 0.03 |
|
|
|
| chr16_+_50355723 | 0.03 |
ENSDART00000166664
|
ENSDARG00000102916
|
ENSDARG00000102916 |
| chr19_-_33013748 | 0.02 |
ENSDART00000134149
|
zgc:91944
|
zgc:91944 |
| chr23_+_25365281 | 0.01 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 2.0 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 2.6 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.2 | 2.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 2.8 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 1.5 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.2 | 4.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 0.5 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.2 | 1.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.2 | 0.5 | GO:0055091 | negative regulation of phospholipase activity(GO:0010519) phospholipid homeostasis(GO:0055091) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.9 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.7 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.1 | 0.3 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.4 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.1 | 1.0 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 0.4 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0046931 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.5 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 1.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.3 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 0.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 2.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.5 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.5 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.4 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.5 | 2.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.5 | 2.8 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.4 | 2.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 0.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.2 | 1.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 2.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.5 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.4 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.7 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 1.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 2.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |