Project
DANIO-CODE
Navigation
Downloads
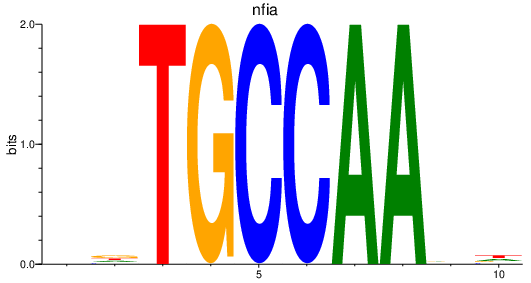
Results for nfia
Z-value: 1.82
Transcription factors associated with nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfia
|
ENSDARG00000062420 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfia | dr10_dc_chr22_-_17026421_17026447 | 0.89 | 4.3e-06 | Click! |
Activity profile of nfia motif
Sorted Z-values of nfia motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfia
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_30673907 | 7.18 |
ENSDART00000053946
|
cxcl12a
|
chemokine (C-X-C motif) ligand 12a (stromal cell-derived factor 1) |
| chr21_-_11539798 | 5.93 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr15_-_29454583 | 5.82 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr12_+_42281025 | 5.72 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr12_-_17590507 | 5.07 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr2_+_2628865 | 4.62 |
ENSDART00000061955
|
myl13
|
myosin, light chain 13 |
| chr5_-_31692803 | 4.62 |
ENSDART00000131983
|
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr9_-_42894582 | 4.58 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr3_+_23090476 | 4.38 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr22_-_27082972 | 4.31 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr3_+_31490043 | 4.27 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr14_-_31676235 | 4.27 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr25_+_19992389 | 3.98 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr9_-_48673183 | 3.83 |
ENSDART00000140185
ENSDART00000134185 |
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr7_-_16776026 | 3.75 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr23_+_31181140 | 3.71 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr5_-_31708933 | 3.70 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr9_-_6949104 | 3.64 |
ENSDART00000059092
|
tmem182a
|
transmembrane protein 182a |
| chr23_+_21737142 | 3.44 |
ENSDART00000066125
|
dhrs3a
|
dehydrogenase/reductase (SDR family) member 3a |
| chr10_+_39141022 | 3.43 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr25_+_33527870 | 3.31 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr14_-_11765407 | 3.15 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr7_-_25623974 | 3.12 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr1_+_1541977 | 3.11 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 4 |
| chr16_+_29716279 | 2.93 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr18_+_23004984 | 2.93 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr25_+_11317025 | 2.85 |
|
|
|
| chr9_-_33584190 | 2.83 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr6_+_14823185 | 2.79 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr9_-_14533551 | 2.73 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr20_-_44598129 | 2.68 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr19_+_12995955 | 2.67 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr18_-_3056732 | 2.67 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr24_+_2523708 | 2.66 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr21_-_23710216 | 2.66 |
ENSDART00000132570
|
ncam1a
|
neural cell adhesion molecule 1a |
| chr9_-_22140954 | 2.62 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr25_+_34135377 | 2.60 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr2_+_38178934 | 2.56 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr25_+_22489716 | 2.55 |
ENSDART00000127831
|
stra6
|
stimulated by retinoic acid 6 |
| chr21_+_21337628 | 2.52 |
ENSDART00000136325
|
rtn2b
|
reticulon 2b |
| chr15_+_7190427 | 2.51 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr17_+_18097490 | 2.50 |
ENSDART00000144894
|
bcl11ba
|
B-cell CLL/lymphoma 11Ba (zinc finger protein) |
| chr16_-_55320569 | 2.49 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr15_-_16271208 | 2.48 |
|
|
|
| chr5_+_31590746 | 2.46 |
|
|
|
| chr23_+_36023748 | 2.40 |
|
|
|
| chr21_-_37286942 | 2.40 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr1_-_11286438 | 2.37 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr14_-_20766448 | 2.33 |
ENSDART00000164146
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr1_+_16683931 | 2.33 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr18_-_39491932 | 2.32 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr15_-_26619404 | 2.32 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr6_-_40774050 | 2.31 |
ENSDART00000076200
|
h1fx
|
H1 histone family, member X |
| chr23_+_20183765 | 2.31 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr7_-_60525749 | 2.29 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr11_+_8558222 | 2.25 |
ENSDART00000169141
ENSDART00000126523 |
tbl1xr1a
|
transducin (beta)-like 1 X-linked receptor 1a |
| chr4_+_15870605 | 2.21 |
ENSDART00000178674
|
BX004864.1
|
ENSDARG00000107641 |
| chr24_-_23893617 | 2.19 |
ENSDART00000080549
|
lyz
|
lysozyme |
| KN150230v1_-_36595 | 2.17 |
|
|
|
| chr23_+_8671759 | 2.17 |
ENSDART00000129997
|
rgs19
|
regulator of G protein signaling 19 |
| chr24_-_1307905 | 2.16 |
ENSDART00000159212
|
nrp1a
|
neuropilin 1a |
| chr7_-_43501747 | 2.16 |
ENSDART00000002279
|
cdh11
|
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr15_-_21901138 | 2.13 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr25_-_16547149 | 2.13 |
ENSDART00000104110
|
slc25a18
|
solute carrier family 25 (glutamate carrier), member 18 |
| chr20_-_20634087 | 2.10 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr3_-_24989269 | 2.09 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr21_+_11410738 | 2.08 |
ENSDART00000146701
|
si:dkey-184p9.7
|
si:dkey-184p9.7 |
| chr3_-_50386208 | 2.06 |
ENSDART00000156709
|
BX088529.1
|
ENSDARG00000097178 |
| chr7_+_23778570 | 2.03 |
ENSDART00000121837
|
efs
|
embryonal Fyn-associated substrate |
| chr9_+_11408481 | 2.03 |
ENSDART00000007308
|
wnt10a
|
wingless-type MMTV integration site family, member 10a |
| chr25_+_14411153 | 2.02 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr15_-_20532537 | 2.00 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr1_-_14642681 | 1.98 |
ENSDART00000109970
|
dlc1
|
DLC1 Rho GTPase activating protein |
| chr16_+_10427961 | 1.96 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr17_-_20877606 | 1.95 |
ENSDART00000088106
|
ank3a
|
ankyrin 3a |
| chr13_+_24703802 | 1.91 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr25_+_30535271 | 1.91 |
|
|
|
| chr14_+_34683602 | 1.91 |
ENSDART00000172171
|
ebf1a
|
early B-cell factor 1a |
| chr17_+_33766838 | 1.89 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr3_+_56008829 | 1.87 |
|
|
|
| chr4_+_4223606 | 1.86 |
|
|
|
| chr2_-_8219329 | 1.86 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr16_-_13704905 | 1.84 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr23_-_28099284 | 1.84 |
ENSDART00000024283
|
sp5l
|
Sp5 transcription factor-like |
| chr23_-_30119058 | 1.84 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr20_+_4222097 | 1.84 |
ENSDART00000097543
|
im:7142702
|
im:7142702 |
| chr1_+_40098861 | 1.79 |
ENSDART00000147497
|
cpz
|
carboxypeptidase Z |
| chr4_-_8610868 | 1.78 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr7_+_34482282 | 1.78 |
|
|
|
| chr11_-_29410649 | 1.76 |
ENSDART00000125753
|
rpl22
|
ribosomal protein L22 |
| chr19_-_20148533 | 1.76 |
ENSDART00000163790
|
evx1
|
even-skipped homeobox 1 |
| chr3_+_28450576 | 1.74 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr7_+_18111890 | 1.74 |
ENSDART00000171606
|
cd248a
|
CD248 molecule, endosialin a |
| chr3_-_54884533 | 1.74 |
ENSDART00000115324
|
hbae1.3
|
hemoglobin, alpha embryonic 1.3 |
| chr20_+_25441689 | 1.73 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr15_+_15580544 | 1.73 |
ENSDART00000016024
|
traf4a
|
tnf receptor-associated factor 4a |
| chr7_-_33558939 | 1.72 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr11_+_6754335 | 1.71 |
ENSDART00000104292
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr1_-_50831155 | 1.70 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr13_+_28997676 | 1.70 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr14_-_6822151 | 1.69 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr7_-_51046010 | 1.68 |
ENSDART00000067647
|
rasl11a
|
RAS-like, family 11, member A |
| chr10_-_21096635 | 1.68 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr8_+_29584750 | 1.67 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr17_+_50622804 | 1.66 |
ENSDART00000049464
|
fermt2
|
fermitin family member 2 |
| chr21_+_40460178 | 1.66 |
ENSDART00000163254
|
coro6
|
coronin 6 |
| chr15_+_27431469 | 1.64 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr22_+_16282153 | 1.64 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr15_+_19717045 | 1.64 |
ENSDART00000138680
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr11_+_30482530 | 1.63 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr18_+_44656323 | 1.63 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr21_-_44109455 | 1.62 |
ENSDART00000044599
|
oatx
|
organic anion transporter X |
| chr15_+_29343644 | 1.62 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr14_-_26406720 | 1.61 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr6_-_6835861 | 1.60 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr16_+_51319421 | 1.60 |
|
|
|
| chr8_+_52802679 | 1.60 |
ENSDART00000162698
|
adgrd2
|
adhesion G protein-coupled receptor D2 |
| chr11_+_41246824 | 1.59 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr5_+_59844830 | 1.59 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr2_+_21067708 | 1.59 |
ENSDART00000148400
ENSDART00000021168 |
rxrga
|
retinoid x receptor, gamma a |
| chr3_+_47735048 | 1.59 |
ENSDART00000083024
|
gpr146
|
G protein-coupled receptor 146 |
| chr9_-_22371512 | 1.59 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr1_-_12584179 | 1.57 |
|
|
|
| chr16_+_32605735 | 1.57 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr9_+_21340251 | 1.57 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr11_-_3846064 | 1.57 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr14_-_2780615 | 1.54 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr1_+_16681778 | 1.53 |
|
|
|
| chr9_+_7609372 | 1.53 |
|
|
|
| chr13_+_22350043 | 1.53 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr9_-_32942783 | 1.52 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr20_-_27412546 | 1.52 |
ENSDART00000148361
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr14_-_32877752 | 1.51 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr24_-_16895179 | 1.50 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr7_+_67227810 | 1.50 |
ENSDART00000163840
|
gcshb
|
glycine cleavage system protein H (aminomethyl carrier), b |
| chr1_+_25974174 | 1.49 |
ENSDART00000152144
|
si:dkey-25o16.2
|
si:dkey-25o16.2 |
| chr25_-_13737344 | 1.49 |
|
|
|
| chr10_-_10906027 | 1.48 |
ENSDART00000122657
|
nrarpa
|
notch-regulated ankyrin repeat protein a |
| chr14_+_45895103 | 1.48 |
|
|
|
| chr1_+_29054033 | 1.46 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr5_-_56984800 | 1.44 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr23_-_4175790 | 1.43 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr2_-_30198789 | 1.43 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr19_-_44223619 | 1.43 |
ENSDART00000160879
|
klhl43
|
kelch-like family member 43 |
| chr7_-_53828910 | 1.42 |
ENSDART00000165499
|
rps17
|
ribosomal protein S17 |
| chr20_-_8431338 | 1.42 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr10_-_36690119 | 1.42 |
ENSDART00000077161
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr3_+_25914076 | 1.41 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr3_+_16979428 | 1.39 |
|
|
|
| chr24_-_25102437 | 1.39 |
|
|
|
| chr24_+_21200975 | 1.37 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr12_+_34631232 | 1.37 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr1_-_22170553 | 1.37 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr10_+_3728747 | 1.37 |
|
|
|
| chr7_+_28877570 | 1.36 |
ENSDART00000170342
|
mtbl
|
metallothionein-B-like |
| chr10_+_33935945 | 1.35 |
|
|
|
| chr5_-_45905518 | 1.35 |
ENSDART00000111589
|
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr12_+_46944395 | 1.35 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr25_+_28329427 | 1.34 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr14_-_2780569 | 1.33 |
ENSDART00000090213
|
ndst1a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1a |
| chr3_-_32190009 | 1.33 |
|
|
|
| chr6_+_251026 | 1.32 |
ENSDART00000042970
|
atf4a
|
activating transcription factor 4a |
| chr13_+_13447540 | 1.32 |
ENSDART00000101673
|
foxi2
|
forkhead box I2 |
| chr23_-_1008307 | 1.31 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr6_-_39311711 | 1.31 |
|
|
|
| chr14_-_2440612 | 1.31 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr16_-_38604326 | 1.31 |
ENSDART00000109124
|
rspo2
|
R-spondin 2 |
| chr7_+_65715260 | 1.30 |
|
|
|
| chr23_-_7892862 | 1.30 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr16_+_5778650 | 1.30 |
ENSDART00000131575
|
zgc:158689
|
zgc:158689 |
| chr15_-_31228685 | 1.30 |
ENSDART00000154254
|
ksr1b
|
kinase suppressor of ras 1b |
| chr16_-_14463682 | 1.29 |
ENSDART00000011224
|
itga10
|
integrin, alpha 10 |
| chr20_+_26803282 | 1.28 |
ENSDART00000077753
|
foxc1b
|
forkhead box C1b |
| chr20_-_8431368 | 1.28 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_+_9228336 | 1.28 |
|
|
|
| chr8_-_4638117 | 1.27 |
ENSDART00000019828
|
zgc:63587
|
zgc:63587 |
| chr24_+_26922969 | 1.26 |
|
|
|
| chr15_+_24064257 | 1.26 |
ENSDART00000156714
|
CR394545.1
|
ENSDARG00000097097 |
| chr2_+_39653315 | 1.25 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr14_-_30467807 | 1.25 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr15_-_12484651 | 1.25 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr23_+_31036558 | 1.25 |
ENSDART00000115417
|
si:ch211-197l9.2
|
si:ch211-197l9.2 |
| chr16_+_21109486 | 1.25 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr17_+_42567327 | 1.24 |
ENSDART00000126087
|
qpct
|
glutaminyl-peptide cyclotransferase |
| chr22_+_18364282 | 1.24 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr24_+_34227202 | 1.24 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr9_+_7609527 | 1.23 |
|
|
|
| chr14_+_34683695 | 1.22 |
ENSDART00000170631
|
ebf1a
|
early B-cell factor 1a |
| chr17_-_23707863 | 1.22 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr17_+_48349119 | 1.21 |
ENSDART00000019804
|
CABZ01043017.1
|
ENSDARG00000001937 |
| chr10_+_36710393 | 1.20 |
|
|
|
| chr21_+_26696853 | 1.20 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr20_+_23643050 | 1.19 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr2_+_27005381 | 1.19 |
|
|
|
| chr18_+_31631789 | 1.18 |
ENSDART00000165660
|
dhx36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr25_+_34545289 | 1.17 |
ENSDART00000126625
|
si:dkey-108k21.10
|
si:dkey-108k21.10 |
| chr6_+_13613459 | 1.16 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr4_+_21319791 | 1.16 |
|
|
|
| chr13_+_22119569 | 1.16 |
ENSDART00000108472
|
synpo2la
|
synaptopodin 2-like a |
| KN150487v1_+_38777 | 1.16 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 14.4 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.9 | 2.6 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.8 | 2.4 | GO:0051701 | interaction with host(GO:0051701) |
| 0.7 | 2.7 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.7 | 3.3 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.7 | 5.9 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 2.4 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.6 | 1.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.6 | 1.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.6 | 3.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.5 | 1.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.5 | 3.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.5 | 1.6 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.5 | 1.5 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.5 | 2.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.5 | 4.4 | GO:0070977 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.5 | 2.8 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.5 | 2.7 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.4 | 1.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.8 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.4 | 1.3 | GO:2000583 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.4 | 1.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.4 | 5.1 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.4 | 2.9 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.4 | 2.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 2.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 1.1 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.4 | 1.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 | 1.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 2.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 | 3.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 2.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 1.7 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.3 | 1.0 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.3 | 1.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.3 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 2.1 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.3 | 6.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 7.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.3 | 1.7 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.3 | 5.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 1.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 4.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 1.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.3 | 1.0 | GO:0071635 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.2 | 0.9 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.2 | 0.9 | GO:0006835 | dicarboxylic acid transport(GO:0006835) |
| 0.2 | 1.1 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.2 | 1.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 0.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 1.3 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.2 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 1.3 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.2 | 0.6 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 1.2 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.2 | 0.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.3 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 0.2 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.2 | 3.1 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 1.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.2 | 1.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.4 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.2 | 1.0 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 1.3 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 1.9 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 1.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.9 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 2.3 | GO:0033338 | medial fin development(GO:0033338) |
| 0.1 | 0.4 | GO:0015835 | peptidoglycan transport(GO:0015835) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.1 | 2.3 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 1.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 2.6 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 3.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 2.3 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.4 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.1 | 6.1 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.1 | 1.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 2.0 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.1 | 2.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 2.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 10.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.8 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.1 | 4.9 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.1 | 0.3 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 2.1 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.2 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 2.6 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.4 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 1.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 6.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 1.6 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 0.5 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 0.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.9 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 2.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 1.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.0 | 0.7 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.3 | GO:0035437 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) maintenance of protein localization in endoplasmic reticulum(GO:0035437) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.7 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.1 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 2.8 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 1.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.7 | GO:2000181 | negative regulation of angiogenesis(GO:0016525) negative regulation of blood vessel morphogenesis(GO:2000181) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.5 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 | 1.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.8 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 6.2 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 0.7 | GO:0051480 | regulation of cytosolic calcium ion concentration(GO:0051480) |
| 0.0 | 0.8 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.9 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.6 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 1.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 0.5 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.6 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 1.3 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.4 | 7.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.7 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 2.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 2.9 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.2 | 0.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 10.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 0.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 1.0 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 7.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 2.0 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 3.0 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 4.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.5 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 3.5 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.4 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 3.8 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.1 | 3.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 3.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 6.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.3 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.4 | GO:0031258 | intercalated disc(GO:0014704) lamellipodium membrane(GO:0031258) |
| 0.1 | 0.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 1.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.0 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 5.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 3.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.7 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 2.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 14.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 11.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.7 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 3.8 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.0 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 11.5 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.9 | 2.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.8 | 3.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.7 | 2.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.7 | 5.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 5.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 3.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.5 | 2.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.5 | 2.6 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 3.9 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.4 | 1.2 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.4 | 2.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.3 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.3 | 1.6 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 3.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 5.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.3 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 3.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 0.8 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.3 | 0.8 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.2 | 1.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 14.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 1.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 2.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 4.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.6 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.2 | 1.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 1.0 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.2 | 0.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.6 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.2 | 0.8 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.2 | 1.9 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.2 | 2.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.0 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 1.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.2 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 2.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.1 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.9 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.4 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 1.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 4.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.3 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 4.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 4.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 8.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 3.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.1 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.5 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.2 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 2.6 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 3.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.3 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 0.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.1 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 4.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 1.9 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.9 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 34.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 4.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 3.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 8.8 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 2.4 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.4 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 6.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 1.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 4.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 3.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 3.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.1 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 2.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 2.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 6.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 1.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 2.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 5.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.1 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 2.2 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.1 | 1.0 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 3.9 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |