Project
DANIO-CODE
Navigation
Downloads
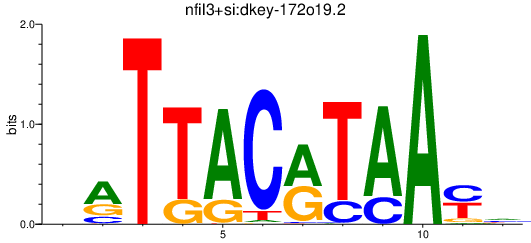
Results for nfil3+si:dkey-172o19.2
Z-value: 0.73
Transcription factors associated with nfil3+si:dkey-172o19.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfil3
|
ENSDARG00000042977 | nuclear factor, interleukin 3 regulated |
|
si_dkey-172o19.2
|
ENSDARG00000071398 | si_dkey-172o19.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfil3 | dr10_dc_chr10_+_5413893_5413957 | -0.55 | 2.9e-02 | Click! |
Activity profile of nfil3+si:dkey-172o19.2 motif
Sorted Z-values of nfil3+si:dkey-172o19.2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfil3+si:dkey-172o19.2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_32747592 | 1.43 |
|
|
|
| chr7_+_44463158 | 1.27 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr9_-_12916555 | 1.25 |
ENSDART00000140691
|
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr12_-_46943717 | 1.20 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr16_+_39209567 | 1.16 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr8_-_37989930 | 1.13 |
|
|
|
| chr8_+_52456064 | 1.13 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr15_-_25025251 | 1.08 |
ENSDART00000109990
|
abhd15a
|
abhydrolase domain containing 15a |
| chr16_-_42116471 | 1.06 |
ENSDART00000058620
|
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr10_+_16914003 | 1.05 |
ENSDART00000177906
|
UNC13B
|
unc-13 homolog B |
| chr7_-_51497945 | 1.00 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr20_-_20921480 | 0.98 |
ENSDART00000152781
|
ckbb
|
creatine kinase, brain b |
| chr23_+_19663746 | 0.97 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr3_-_29779725 | 0.96 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr2_+_3115593 | 0.95 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr23_+_540624 | 0.95 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr22_+_4145005 | 0.94 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr21_+_43183522 | 0.93 |
ENSDART00000151350
|
aff4
|
AF4/FMR2 family, member 4 |
| chr16_-_29452509 | 0.90 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr25_+_34340569 | 0.90 |
ENSDART00000157638
|
tmem231
|
transmembrane protein 231 |
| chr24_+_16838877 | 0.90 |
ENSDART00000145520
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr16_+_27680365 | 0.88 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr20_-_44678938 | 0.87 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr19_+_15536640 | 0.86 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr10_+_18953503 | 0.85 |
ENSDART00000030205
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr13_+_33578737 | 0.85 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr19_-_868378 | 0.84 |
|
|
|
| chr19_-_35411993 | 0.82 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr5_+_22470117 | 0.82 |
ENSDART00000020434
|
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr24_-_23639325 | 0.82 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr8_-_53165501 | 0.79 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr7_+_17848688 | 0.78 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr17_-_17739301 | 0.75 |
ENSDART00000157128
|
adck1
|
aarF domain containing kinase 1 |
| chr7_+_71263100 | 0.75 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr16_+_41004372 | 0.75 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr10_+_6925373 | 0.75 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr14_+_3399943 | 0.75 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr8_-_410190 | 0.74 |
ENSDART00000151155
|
trim36
|
tripartite motif containing 36 |
| chr8_+_23806179 | 0.74 |
ENSDART00000040362
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr18_+_5616001 | 0.73 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr12_-_42212994 | 0.72 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr23_-_1562572 | 0.71 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr25_-_19889225 | 0.70 |
|
|
|
| chr2_+_35621136 | 0.70 |
ENSDART00000133018
ENSDART00000147278 |
plk3
|
polo-like kinase 3 (Drosophila) |
| chr14_-_745312 | 0.70 |
ENSDART00000049918
ENSDART00000165856 |
trim35-27
|
tripartite motif containing 35-27 |
| chr7_+_71262845 | 0.68 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr13_+_33331767 | 0.68 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr5_+_29231012 | 0.68 |
ENSDART00000159327
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr19_+_14592632 | 0.67 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr15_+_36075206 | 0.67 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr4_-_71601845 | 0.67 |
ENSDART00000174158
|
CABZ01021433.1
|
ENSDARG00000105696 |
| chr14_+_29945327 | 0.66 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr7_+_45747622 | 0.66 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr2_-_6380292 | 0.66 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr17_-_36913213 | 0.65 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr9_+_22846286 | 0.65 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr7_-_37283707 | 0.64 |
ENSDART00000148905
ENSDART00000150229 |
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr5_+_29231241 | 0.64 |
ENSDART00000131687
ENSDART00000035098 |
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr8_-_29842489 | 0.64 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr14_-_36101782 | 0.63 |
|
|
|
| chr3_-_29779598 | 0.63 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr5_-_23211957 | 0.63 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr10_+_19636552 | 0.63 |
|
|
|
| chr18_-_7522421 | 0.63 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr15_+_24802510 | 0.63 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr11_+_24583090 | 0.62 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr24_+_37450437 | 0.62 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr24_-_37450413 | 0.62 |
ENSDART00000056303
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr10_+_27028355 | 0.62 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr1_+_23685562 | 0.61 |
ENSDART00000102542
ENSDART00000160882 |
ENSDARG00000090822
qdprb2
|
ENSDARG00000090822 quinoid dihydropteridine reductase b2 |
| chr19_-_44459352 | 0.61 |
ENSDART00000151084
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr21_-_43641419 | 0.60 |
ENSDART00000151486
ENSDART00000151115 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr15_+_24741931 | 0.60 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr24_+_16838984 | 0.60 |
ENSDART00000023833
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr4_-_14208573 | 0.60 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr25_+_21001126 | 0.60 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr15_+_15296839 | 0.60 |
ENSDART00000033904
|
med29
|
mediator complex subunit 29 |
| chr2_+_46210971 | 0.60 |
|
|
|
| chr11_+_38858311 | 0.59 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr17_-_2359241 | 0.59 |
ENSDART00000155227
|
si:dkey-248g15.2
|
si:dkey-248g15.2 |
| chr3_-_40142513 | 0.59 |
ENSDART00000018626
|
pdap1b
|
pdgfa associated protein 1b |
| chr5_+_22470177 | 0.59 |
ENSDART00000020434
|
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr25_-_13394261 | 0.59 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr3_-_26052785 | 0.59 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr2_-_3115484 | 0.59 |
|
|
|
| chr10_+_17277353 | 0.58 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr23_-_29886117 | 0.58 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr16_-_39209426 | 0.58 |
|
|
|
| chr7_+_73447957 | 0.58 |
ENSDART00000050357
ENSDART00000110769 |
btr12
|
bloodthirsty-related gene family, member 12 |
| chr6_+_44199738 | 0.57 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr4_-_14916416 | 0.57 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr1_-_30299017 | 0.57 |
|
|
|
| chr1_+_33962885 | 0.57 |
ENSDART00000145535
|
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr10_+_18953548 | 0.57 |
ENSDART00000030205
|
bnip3lb
|
BCL2/adenovirus E1B interacting protein 3-like b |
| chr17_+_51655501 | 0.56 |
ENSDART00000149807
|
odc1
|
ornithine decarboxylase 1 |
| chr1_-_51075777 | 0.56 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr15_+_5125138 | 0.56 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr5_-_32636372 | 0.56 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr19_-_26188624 | 0.56 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr3_-_26052601 | 0.56 |
ENSDART00000147517
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr5_+_24016742 | 0.55 |
ENSDART00000144560
|
dgcr8
|
DGCR8 microprocessor complex subunit |
| chr21_+_25764848 | 0.55 |
ENSDART00000136146
ENSDART00000035062 |
nf2b
|
neurofibromin 2b (merlin) |
| chr20_+_54404987 | 0.54 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr2_+_55112111 | 0.54 |
ENSDART00000163834
|
rnf126
|
ring finger protein 126 |
| chr7_+_16771283 | 0.54 |
ENSDART00000171275
|
CU929054.1
|
ENSDARG00000102605 |
| chr24_-_23639458 | 0.54 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr15_+_24741620 | 0.54 |
ENSDART00000078014
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr13_-_50940551 | 0.54 |
|
|
|
| chr22_-_5600910 | 0.54 |
ENSDART00000179579
|
CABZ01075081.1
|
ENSDARG00000106226 |
| chr14_-_30564987 | 0.53 |
ENSDART00000173451
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr11_-_15886860 | 0.53 |
ENSDART00000170731
|
zgc:173544
|
zgc:173544 |
| chr16_+_41004516 | 0.53 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr19_+_27338879 | 0.53 |
ENSDART00000149988
|
nelfe
|
negative elongation factor complex member E |
| chr13_+_46652067 | 0.53 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr21_-_4842701 | 0.53 |
ENSDART00000146766
|
rnf165a
|
ring finger protein 165a |
| chr13_-_36486255 | 0.53 |
ENSDART00000160250
|
BX927407.1
|
ENSDARG00000103360 |
| chr25_-_20932941 | 0.53 |
ENSDART00000138985
ENSDART00000046298 |
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr23_-_29952020 | 0.52 |
ENSDART00000058407
|
slc25a33
|
solute carrier family 25 (pyrimidine nucleotide carrier), member 33 |
| chr23_-_25208472 | 0.52 |
ENSDART00000103989
ENSDART00000160278 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr25_+_22183891 | 0.52 |
ENSDART00000089516
|
stoml1
|
stomatin (EPB72)-like 1 |
| chr3_-_26060787 | 0.52 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr16_+_47283253 | 0.52 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr9_-_30448622 | 0.52 |
ENSDART00000129926
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr16_+_28946580 | 0.52 |
ENSDART00000146525
|
chtopb
|
chromatin target of PRMT1b |
| chr14_+_47040687 | 0.52 |
ENSDART00000113469
|
nocta
|
nocturnin a |
| chr5_-_48028931 | 0.52 |
ENSDART00000031194
|
lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr24_-_19573615 | 0.52 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr3_+_40142679 | 0.52 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr15_-_25025058 | 0.51 |
ENSDART00000109990
|
abhd15a
|
abhydrolase domain containing 15a |
| chr2_-_40067309 | 0.51 |
|
|
|
| chr15_+_29092022 | 0.51 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr8_+_41003546 | 0.51 |
ENSDART00000129344
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr10_+_7634671 | 0.51 |
ENSDART00000171744
|
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_+_46474561 | 0.51 |
ENSDART00000167051
|
cbr1
|
carbonyl reductase 1 |
| chr16_-_44979403 | 0.51 |
|
|
|
| chr2_-_17443607 | 0.51 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr17_+_25168837 | 0.50 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr6_-_32424559 | 0.50 |
ENSDART00000151002
|
usp1
|
ubiquitin specific peptidase 1 |
| chr15_-_16137131 | 0.50 |
|
|
|
| chr13_-_9034919 | 0.50 |
ENSDART00000135373
|
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr5_+_26888744 | 0.50 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr16_+_25202230 | 0.49 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr12_+_20569659 | 0.49 |
ENSDART00000141804
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr17_+_27988842 | 0.49 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr11_+_13118866 | 0.49 |
ENSDART00000123257
|
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr8_+_21101133 | 0.49 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr2_+_3115688 | 0.49 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr9_+_19812730 | 0.48 |
ENSDART00000110009
|
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr9_-_46614763 | 0.48 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr2_+_46210909 | 0.48 |
|
|
|
| chr5_-_68751287 | 0.48 |
ENSDART00000112692
|
ENSDARG00000077155
|
ENSDARG00000077155 |
| chr1_+_48770997 | 0.48 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_-_17443642 | 0.48 |
ENSDART00000136207
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr3_+_19095975 | 0.48 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr13_+_3807780 | 0.48 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr21_-_32027717 | 0.47 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr23_-_32239909 | 0.47 |
|
|
|
| chr13_+_29904945 | 0.47 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr21_-_32747544 | 0.47 |
|
|
|
| chr19_-_868837 | 0.47 |
|
|
|
| chr17_-_22552999 | 0.47 |
ENSDART00000141523
|
exo1
|
exonuclease 1 |
| chr6_+_50452131 | 0.47 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr17_+_13078410 | 0.47 |
ENSDART00000149779
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr16_+_40181425 | 0.47 |
ENSDART00000155421
|
cenpw
|
centromere protein W |
| chr12_+_34157065 | 0.46 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr21_-_39521698 | 0.46 |
ENSDART00000020174
|
dynll2b
|
dynein, light chain, LC8-type 2b |
| chr19_+_43984165 | 0.46 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr11_-_44107913 | 0.46 |
|
|
|
| chr19_-_44161951 | 0.46 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr24_-_6647047 | 0.46 |
|
|
|
| chr12_-_28679813 | 0.46 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr2_+_10858480 | 0.46 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr25_-_36616544 | 0.45 |
|
|
|
| chr5_-_47610384 | 0.45 |
|
|
|
| chr17_+_25815708 | 0.45 |
ENSDART00000164160
|
acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr16_+_47283374 | 0.45 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr19_+_14592246 | 0.45 |
ENSDART00000161088
ENSDART00000161965 |
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr17_+_16082983 | 0.45 |
ENSDART00000133154
|
znf395a
|
zinc finger protein 395a |
| chr5_+_56988663 | 0.45 |
ENSDART00000074268
|
zgc:153929
|
zgc:153929 |
| chr22_+_16070849 | 0.45 |
ENSDART00000165216
|
dph5
|
diphthamide biosynthesis 5 |
| KN150266v1_-_68652 | 0.45 |
|
|
|
| chr1_+_34763140 | 0.45 |
ENSDART00000053806
|
gab1
|
GRB2-associated binding protein 1 |
| chr12_-_10327938 | 0.45 |
ENSDART00000007335
|
ndc80
|
NDC80 kinetochore complex component |
| chr3_-_29779552 | 0.45 |
ENSDART00000151501
|
RUNDC1
|
RUN domain containing 1 |
| chr10_+_1653720 | 0.44 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr11_+_16017857 | 0.44 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr2_+_30800532 | 0.44 |
|
|
|
| chr13_+_15685745 | 0.44 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr3_+_37507517 | 0.44 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| KN150030v1_-_22572 | 0.44 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr19_-_17306473 | 0.44 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr20_-_3301951 | 0.44 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr1_+_34763197 | 0.44 |
ENSDART00000053806
|
gab1
|
GRB2-associated binding protein 1 |
| chr7_+_28341426 | 0.44 |
ENSDART00000019991
|
slc7a6os
|
solute carrier family 7, member 6 opposite strand |
| chr22_+_21373501 | 0.44 |
ENSDART00000089408
|
shdb
|
Src homology 2 domain containing transforming protein D, b |
| chr21_+_18870471 | 0.44 |
ENSDART00000160185
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr15_+_5124690 | 0.44 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr8_-_410293 | 0.43 |
ENSDART00000164886
|
trim36
|
tripartite motif containing 36 |
| chr24_+_21531899 | 0.43 |
ENSDART00000158833
|
si:ch211-140b10.6
|
si:ch211-140b10.6 |
| chr19_-_48327666 | 0.43 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0032516 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.5 | 1.4 | GO:0045822 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.3 | 1.4 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.3 | 1.0 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.3 | 0.9 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.2 | 0.7 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 0.2 | GO:0043470 | regulation of carbohydrate catabolic process(GO:0043470) regulation of cellular carbohydrate catabolic process(GO:0043471) |
| 0.2 | 0.7 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 1.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.2 | 2.0 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.2 | 0.6 | GO:0046824 | snRNA export from nucleus(GO:0006408) regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 1.3 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.7 | GO:1901741 | positive regulation of syncytium formation by plasma membrane fusion(GO:0060143) positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 1.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.2 | 1.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.5 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.2 | 0.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 1.8 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.1 | 1.8 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.8 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.5 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.5 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.4 | GO:2000242 | negative regulation of reproductive process(GO:2000242) |
| 0.1 | 0.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.3 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 0.6 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.3 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.5 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.4 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.8 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.6 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.3 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.8 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 0.6 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 1.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.4 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.1 | 1.1 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.2 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.8 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.4 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.7 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.4 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 1.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.4 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.9 | GO:0042559 | pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.1 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.1 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.2 | GO:0042126 | nitrate metabolic process(GO:0042126) nitrate assimilation(GO:0042128) |
| 0.1 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.5 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.2 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.2 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.3 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.8 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.0 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.5 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.0 | GO:0032885 | regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.0 | 0.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.9 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 1.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.5 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.3 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 1.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 1.5 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0032635 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0010481 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0043649 | coenzyme catabolic process(GO:0009109) dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 1.3 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.8 | GO:0006458 | 'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.0 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.3 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.9 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.6 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.9 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.3 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.0 | 0.4 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.1 | GO:0030730 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.6 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.5 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 3.8 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) negative regulation of DNA-dependent DNA replication(GO:2000104) |
| 0.0 | 0.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:0038066 | p38MAPK cascade(GO:0038066) regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 0.4 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.4 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0061462 | protein localization to lysosome(GO:0061462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.2 | 1.4 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 0.8 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 3.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 1.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.4 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.8 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.5 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.9 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 1.4 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.3 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 2.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0044304 | main axon(GO:0044304) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:1990077 | primosome complex(GO:1990077) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.8 | GO:0098573 | intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.3 | 0.9 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.2 | 0.9 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 0.6 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.2 | 1.0 | GO:0052834 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.2 | 0.7 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.2 | 1.0 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 1.0 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.5 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 0.7 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 0.5 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.1 | 1.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.5 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.1 | 0.5 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.1 | 0.7 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.1 | 0.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 0.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 0.8 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.6 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.8 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.9 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 1.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.3 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.1 | 0.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) double-strand/single-strand DNA junction binding(GO:0000406) flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.4 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.1 | 0.6 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 1.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0046857 | nitrite reductase [NAD(P)H] activity(GO:0008942) oxidoreductase activity, acting on other nitrogenous compounds as donors, with NAD or NADP as acceptor(GO:0046857) |
| 0.1 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.3 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.6 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 1.8 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.2 | GO:0042979 | ornithine decarboxylase inhibitor activity(GO:0008073) ornithine decarboxylase regulator activity(GO:0042979) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.4 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 2.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.5 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0070300 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.8 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0016894 | endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 3'-phosphomonoesters(GO:0016894) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.1 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.0 | GO:0052657 | guanine phosphoribosyltransferase activity(GO:0052657) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.0 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 0.9 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 0.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.9 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.4 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 0.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.0 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.6 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 1.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.5 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |