Project
DANIO-CODE
Navigation
Downloads
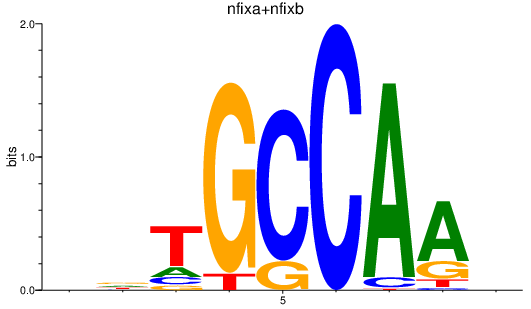
Results for nfixa+nfixb
Z-value: 0.81
Transcription factors associated with nfixa+nfixb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfixa
|
ENSDARG00000043226 | nuclear factor I/Xa |
|
nfixb
|
ENSDARG00000061836 | nuclear factor I/Xb |
Activity profile of nfixa+nfixb motif
Sorted Z-values of nfixa+nfixb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfixa+nfixb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_36503296 | 2.19 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr6_+_28218420 | 1.78 |
ENSDART00000171216
ENSDART00000171377 |
si:ch73-14h10.2
|
si:ch73-14h10.2 |
| chr22_-_8144696 | 1.70 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr21_-_32747592 | 1.69 |
|
|
|
| chr23_+_19663746 | 1.65 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr24_+_31751950 | 1.59 |
ENSDART00000156493
|
CT737162.1
|
ENSDARG00000096998 |
| chr3_+_32285237 | 1.55 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr22_-_8144599 | 1.54 |
ENSDART00000156571
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr5_-_23211957 | 1.44 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr23_-_27579257 | 1.35 |
ENSDART00000137229
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr22_+_25113293 | 1.33 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr7_+_28453707 | 1.32 |
ENSDART00000173947
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr12_+_21176669 | 1.32 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr23_-_22596648 | 1.28 |
ENSDART00000079019
|
spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr23_+_25062876 | 1.26 |
ENSDART00000145307
ENSDART00000172299 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr1_-_54586004 | 1.23 |
ENSDART00000152687
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr13_-_226574 | 1.14 |
|
|
|
| chr2_-_45810787 | 1.09 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr25_+_16098033 | 1.07 |
ENSDART00000141994
|
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr12_-_3042394 | 1.07 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr21_+_15608741 | 1.06 |
ENSDART00000146909
|
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr23_+_25062826 | 1.06 |
ENSDART00000172299
|
arhgap4a
|
Rho GTPase activating protein 4a |
| chr16_+_27680365 | 1.01 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr14_+_30434569 | 1.00 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr12_-_17302222 | 0.94 |
ENSDART00000079144
|
ptenb
|
phosphatase and tensin homolog B |
| chr24_+_19270877 | 0.94 |
|
|
|
| chr17_-_23201467 | 0.93 |
ENSDART00000156411
|
fam98a
|
family with sequence similarity 98, member A |
| chr7_+_58910235 | 0.92 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr23_+_16904665 | 0.90 |
ENSDART00000104782
|
zgc:173570
|
zgc:173570 |
| chr15_+_17407036 | 0.83 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr6_-_52567422 | 0.83 |
ENSDART00000098421
|
uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr19_+_32000438 | 0.81 |
ENSDART00000150910
|
gmnn
|
geminin, DNA replication inhibitor |
| chr15_-_17088654 | 0.77 |
ENSDART00000154719
ENSDART00000014465 |
hip1
|
huntingtin interacting protein 1 |
| chr23_+_6610443 | 0.77 |
ENSDART00000005373
|
spo11
|
SPO11 meiotic protein covalently bound to DSB |
| chr4_+_17666992 | 0.76 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr24_+_32497012 | 0.75 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr9_-_2522639 | 0.74 |
ENSDART00000137706
|
scrn3
|
secernin 3 |
| chr6_-_9046536 | 0.73 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr3_-_55399158 | 0.73 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr3_+_32530476 | 0.73 |
ENSDART00000171556
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr20_-_42344217 | 0.73 |
ENSDART00000034054
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr19_-_3547196 | 0.71 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr16_-_13723778 | 0.70 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr11_-_13094557 | 0.70 |
ENSDART00000160989
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr9_+_3577477 | 0.69 |
ENSDART00000147650
|
tlk1a
|
tousled-like kinase 1a |
| chr20_+_29788065 | 0.67 |
ENSDART00000164121
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr18_+_20479090 | 0.65 |
ENSDART00000100665
ENSDART00000147867 |
ddb2
|
damage-specific DNA binding protein 2 |
| chr5_-_29782945 | 0.65 |
ENSDART00000125381
|
gig2o
|
grass carp reovirus (GCRV)-induced gene 2o |
| chr6_-_41141242 | 0.64 |
ENSDART00000128723
ENSDART00000151055 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr21_+_15608551 | 0.62 |
ENSDART00000024858
|
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr6_-_41141280 | 0.62 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr20_+_31375056 | 0.59 |
|
|
|
| chr3_-_29760674 | 0.59 |
ENSDART00000014021
|
slc25a39
|
solute carrier family 25, member 39 |
| chr10_-_22180530 | 0.58 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr1_-_54585975 | 0.57 |
ENSDART00000152504
|
si:ch211-286b5.4
|
si:ch211-286b5.4 |
| chr13_+_969851 | 0.56 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr2_-_45810714 | 0.56 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr10_+_43953171 | 0.55 |
|
|
|
| chr9_+_27518890 | 0.55 |
ENSDART00000111039
|
GTPBP8
|
GTP binding protein 8 (putative) |
| chr1_+_40428722 | 0.54 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr3_+_35140906 | 0.54 |
|
|
|
| chr17_+_25545565 | 0.53 |
ENSDART00000121700
|
qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr5_-_34664282 | 0.53 |
ENSDART00000114981
ENSDART00000051279 |
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr12_-_17302254 | 0.53 |
ENSDART00000079138
|
ptenb
|
phosphatase and tensin homolog B |
| chr20_-_38851375 | 0.52 |
|
|
|
| chr22_-_5791916 | 0.52 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr17_-_33754959 | 0.52 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr11_-_11591523 | 0.51 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr24_+_15510787 | 0.51 |
ENSDART00000042943
|
fbxo15
|
F-box protein 15 |
| chr16_-_29517367 | 0.51 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr1_+_18390488 | 0.51 |
ENSDART00000002886
ENSDART00000158745 |
exosc9
|
exosome component 9 |
| chr18_+_5066229 | 0.50 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr8_-_18849529 | 0.50 |
ENSDART00000100491
|
sh3gl1b
|
SH3-domain GRB2-like 1b |
| chr14_+_47368995 | 0.49 |
ENSDART00000060577
|
tmem33
|
transmembrane protein 33 |
| chr13_+_39151166 | 0.48 |
ENSDART00000113259
|
ENSDARG00000079055
|
ENSDARG00000079055 |
| chr5_-_36503236 | 0.47 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr8_-_20106132 | 0.46 |
ENSDART00000029939
|
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr14_+_28167358 | 0.44 |
ENSDART00000157306
ENSDART00000166375 ENSDART00000167439 ENSDART00000017075 |
xiap
|
X-linked inhibitor of apoptosis |
| chr17_-_14443647 | 0.44 |
ENSDART00000039438
|
jkamp
|
jnk1/mapk8-associated membrane protein |
| chr25_+_15901398 | 0.44 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr8_+_19945820 | 0.44 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr12_-_3042573 | 0.44 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr7_-_73620443 | 0.43 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr14_-_21779536 | 0.43 |
|
|
|
| chr9_+_9956221 | 0.43 |
ENSDART00000168559
|
ttf2
|
transcription termination factor, RNA polymerase II |
| chr16_-_22397159 | 0.43 |
|
|
|
| chr22_+_25156826 | 0.43 |
ENSDART00000140670
ENSDART00000105308 |
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr12_-_36282538 | 0.41 |
ENSDART00000165935
|
nploc4
|
NPL4 homolog, ubiquitin recognition factor |
| chr15_+_44909356 | 0.40 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr5_-_24013767 | 0.40 |
ENSDART00000003957
|
tia1l
|
cytotoxic granule-associated RNA binding protein 1, like |
| chr14_+_28132910 | 0.40 |
ENSDART00000006489
|
acsl4a
|
acyl-CoA synthetase long-chain family member 4a |
| chr21_-_32747544 | 0.40 |
|
|
|
| chr8_+_21127033 | 0.37 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr13_+_35799851 | 0.37 |
ENSDART00000046115
|
mfsd2aa
|
major facilitator superfamily domain containing 2aa |
| chr10_-_35164761 | 0.37 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr22_-_5791865 | 0.36 |
ENSDART00000011076
|
cers5
|
ceramide synthase 5 |
| chr3_-_15861110 | 0.35 |
ENSDART00000080672
|
mrps34
|
mitochondrial ribosomal protein S34 |
| chr25_-_29986604 | 0.35 |
ENSDART00000176535
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr3_-_40791555 | 0.34 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr23_-_25208472 | 0.34 |
ENSDART00000103989
ENSDART00000160278 |
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr3_+_30956652 | 0.34 |
ENSDART00000129564
|
si:dkey-66i24.8
|
si:dkey-66i24.8 |
| chr10_+_24452609 | 0.33 |
ENSDART00000140572
ENSDART00000139850 ENSDART00000109663 |
PAXBP1
|
PAX3 and PAX7 binding protein 1 |
| chr20_-_54160322 | 0.32 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr24_+_21030422 | 0.31 |
ENSDART00000154940
|
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr13_-_10488076 | 0.30 |
ENSDART00000160561
|
si:ch73-54n14.2
|
si:ch73-54n14.2 |
| chr17_+_14776302 | 0.30 |
ENSDART00000154229
|
RTRAF
|
zgc:56576 |
| chr20_+_3573418 | 0.30 |
|
|
|
| chr14_-_26113593 | 0.30 |
ENSDART00000020582
|
tmed9
|
transmembrane p24 trafficking protein 9 |
| chr21_+_33216833 | 0.29 |
|
|
|
| chr11_-_11906170 | 0.29 |
ENSDART00000081544
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr13_+_21637976 | 0.29 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| KN149787v1_-_20460 | 0.29 |
|
|
|
| chr12_+_44772179 | 0.29 |
ENSDART00000112750
|
dhx32a
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32a |
| chr7_-_24778185 | 0.28 |
ENSDART00000135179
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr12_-_33687845 | 0.28 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr18_-_36780732 | 0.27 |
|
|
|
| chr3_+_30956719 | 0.27 |
ENSDART00000129564
|
si:dkey-66i24.8
|
si:dkey-66i24.8 |
| chr10_+_38981942 | 0.27 |
|
|
|
| chr23_-_15090815 | 0.27 |
ENSDART00000147636
|
ndrg3b
|
ndrg family member 3b |
| chr23_+_5661715 | 0.27 |
ENSDART00000164400
|
si:ch1073-182o11.2
|
si:ch1073-182o11.2 |
| chr6_+_49489473 | 0.26 |
ENSDART00000177749
|
FO704848.1
|
ENSDARG00000107438 |
| chr11_-_13094160 | 0.26 |
ENSDART00000161983
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr20_-_33887129 | 0.26 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr19_+_14489756 | 0.25 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr20_+_31375136 | 0.25 |
|
|
|
| chr2_-_6545431 | 0.25 |
ENSDART00000161934
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr3_+_43124830 | 0.25 |
ENSDART00000157619
|
litaf
|
lipopolysaccharide-induced TNF factor |
| chr23_+_36494483 | 0.23 |
ENSDART00000042701
|
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr24_+_22614280 | 0.23 |
ENSDART00000135392
|
si:dkey-7n6.2
|
si:dkey-7n6.2 |
| chr7_-_69114652 | 0.23 |
|
|
|
| chr3_-_36460498 | 0.22 |
ENSDART00000161501
|
rrn3
|
RRN3 homolog, RNA polymerase I transcription factor |
| chr9_+_3577705 | 0.22 |
|
|
|
| chr21_-_22362339 | 0.21 |
ENSDART00000138128
|
si:ch73-112l6.1
|
si:ch73-112l6.1 |
| chr8_-_11732278 | 0.19 |
ENSDART00000172431
|
anapc7
|
anaphase promoting complex subunit 7 |
| chr8_-_31044580 | 0.19 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr5_-_31708973 | 0.18 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr3_+_32285564 | 0.17 |
ENSDART00000156986
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr7_-_24778134 | 0.17 |
ENSDART00000135179
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr5_-_51184313 | 0.17 |
ENSDART00000159166
|
homer1b
|
homer scaffolding protein 1b |
| chr9_+_54736764 | 0.16 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr1_+_35919158 | 0.16 |
ENSDART00000176692
|
CR762429.1
|
ENSDARG00000108193 |
| chr3_-_55399071 | 0.16 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr21_-_9689702 | 0.16 |
ENSDART00000158836
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr7_-_51373670 | 0.15 |
ENSDART00000109055
|
cited1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr3_+_31468838 | 0.15 |
ENSDART00000076640
|
ccdc43
|
coiled-coil domain containing 43 |
| chr15_-_19398506 | 0.15 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr6_-_19813235 | 0.15 |
ENSDART00000151152
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr12_-_36415669 | 0.15 |
|
|
|
| chr25_+_26962541 | 0.14 |
ENSDART00000115139
|
pot1
|
protection of telomeres 1 homolog |
| chr11_-_11906118 | 0.14 |
ENSDART00000081544
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr15_-_19398601 | 0.14 |
ENSDART00000062576
|
thyn1
|
thymocyte nuclear protein 1 |
| chr21_+_35006904 | 0.14 |
ENSDART00000021645
|
kdelc2
|
KDEL (Lys-Asp-Glu-Leu) containing 2 |
| chr14_-_6822444 | 0.14 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr10_-_28474052 | 0.14 |
|
|
|
| chr15_-_8215797 | 0.13 |
ENSDART00000156663
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr22_-_15576405 | 0.13 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr5_-_37281096 | 0.12 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr9_+_6609374 | 0.12 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr1_-_32825385 | 0.11 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr2_-_45810868 | 0.11 |
ENSDART00000135665
ENSDART00000075080 |
prpf38b
|
pre-mRNA processing factor 38B |
| chr8_-_20105940 | 0.10 |
ENSDART00000133141
|
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr5_-_32860888 | 0.10 |
ENSDART00000085636
|
ENSDARG00000060631
|
ENSDARG00000060631 |
| chr16_-_10625997 | 0.10 |
ENSDART00000110606
|
abhd16a
|
abhydrolase domain containing 16A |
| chr17_+_23202235 | 0.09 |
ENSDART00000164708
ENSDART00000064993 |
si:ch211-195b21.5
|
si:ch211-195b21.5 |
| chr5_+_52362499 | 0.09 |
ENSDART00000157836
|
hint2
|
histidine triad nucleotide binding protein 2 |
| chr6_+_54857805 | 0.09 |
|
|
|
| chr14_-_2440612 | 0.08 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr13_-_26573327 | 0.08 |
ENSDART00000141232
|
BX005477.1
|
ENSDARG00000093327 |
| chr19_+_25880730 | 0.08 |
ENSDART00000018553
|
rpa3
|
replication protein A3 |
| chr25_+_15901561 | 0.07 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr9_+_24315161 | 0.06 |
ENSDART00000090346
|
pgap1
|
post-GPI attachment to proteins 1 |
| chr21_-_33196706 | 0.06 |
ENSDART00000003983
|
rbm22
|
RNA binding motif protein 22 |
| chr23_+_36494057 | 0.06 |
ENSDART00000042701
|
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr3_-_22098046 | 0.05 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr13_-_30807601 | 0.05 |
|
|
|
| chr18_+_27018924 | 0.04 |
ENSDART00000133547
|
pik3c2a
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr22_+_9493668 | 0.04 |
ENSDART00000143953
|
strip1
|
striatin interacting protein 1 |
| chr4_+_40092288 | 0.04 |
|
|
|
| chr13_-_4850183 | 0.03 |
ENSDART00000132931
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr9_-_22371512 | 0.03 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr8_+_27536121 | 0.03 |
ENSDART00000016696
|
rhocb
|
ras homolog family member Cb |
| chr16_+_46463904 | 0.02 |
|
|
|
| chr13_-_46424420 | 0.02 |
|
|
|
| chr14_+_21779325 | 0.01 |
ENSDART00000106147
|
slc43a1a
|
solute carrier family 43 (amino acid system L transporter), member 1a |
| chr12_+_36415304 | 0.00 |
ENSDART00000164192
ENSDART00000166309 ENSDART00000175409 |
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr22_+_11827925 | 0.00 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr9_+_2346341 | 0.00 |
ENSDART00000135180
ENSDART00000062292 |
atf2
|
activating transcription factor 2 |
| chr5_+_44296157 | 0.00 |
ENSDART00000145624
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.3 | 0.8 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.2 | 0.7 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.2 | 1.5 | GO:0035284 | fourth ventricle development(GO:0021592) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.4 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.5 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.5 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 | 0.6 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.5 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.8 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:1990379 | maintenance of blood-brain barrier(GO:0035633) lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.1 | 0.4 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.5 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.3 | GO:2000737 | negative regulation of stem cell differentiation(GO:2000737) |
| 0.1 | 0.2 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.1 | 0.8 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.6 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.9 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.3 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.9 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.0 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.6 | GO:0009411 | response to UV(GO:0009411) |
| 0.0 | 1.1 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.7 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 0.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 0.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 1.8 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 2.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 2.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.4 | 1.5 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 0.7 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.8 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.4 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.3 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 1.0 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.1 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.9 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 2.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |