Project
DANIO-CODE
Navigation
Downloads
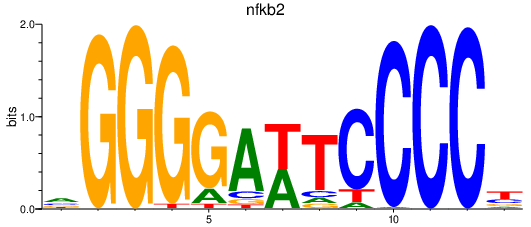
Results for nfkb2
Z-value: 0.73
Transcription factors associated with nfkb2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nfkb2
|
ENSDARG00000038687 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nfkb2 | dr10_dc_chr13_+_22589282_22589368 | 0.64 | 7.3e-03 | Click! |
Activity profile of nfkb2 motif
Sorted Z-values of nfkb2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nfkb2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_28680035 | 2.20 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr6_-_8468819 | 2.10 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr6_+_3174047 | 1.73 |
ENSDART00000160290
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr6_-_8468961 | 1.64 |
ENSDART00000064149
|
nabp1b
|
nucleic acid binding protein 1b |
| chr3_-_31487744 | 1.56 |
ENSDART00000124559
|
moto
|
minamoto |
| chr9_+_188749 | 1.40 |
ENSDART00000172946
|
rrp1
|
ribosomal RNA processing 1 |
| chr12_-_28679813 | 1.38 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr6_+_38629054 | 1.28 |
ENSDART00000086533
|
atp10a
|
ATPase, class V, type 10A |
| chr19_-_3547196 | 1.23 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr13_+_36832090 | 1.19 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr5_+_23582676 | 1.17 |
ENSDART00000139520
|
tp53
|
tumor protein p53 |
| chr12_-_28680442 | 1.15 |
ENSDART00000178777
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr20_+_39145226 | 1.08 |
ENSDART00000142778
|
BX569784.1
|
ENSDARG00000092980 |
| chr21_-_23873574 | 0.97 |
|
|
|
| chr5_+_23582805 | 0.96 |
ENSDART00000137298
|
tp53
|
tumor protein p53 |
| chr1_+_48770997 | 0.92 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr9_-_20743808 | 0.89 |
|
|
|
| chr23_+_25952958 | 0.87 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr21_+_36938203 | 0.85 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr21_+_36938230 | 0.84 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr8_-_40149223 | 0.84 |
ENSDART00000005118
|
gpx8
|
glutathione peroxidase 8 (putative) |
| chr8_-_12394688 | 0.83 |
ENSDART00000133350
|
traf1
|
TNF receptor-associated factor 1 |
| chr5_-_31089598 | 0.80 |
ENSDART00000098172
|
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
| chr10_+_21487458 | 0.77 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr14_+_27984076 | 0.75 |
ENSDART00000173237
|
mid2
|
midline 2 |
| chr9_+_20743219 | 0.71 |
ENSDART00000147435
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr9_+_20743866 | 0.65 |
ENSDART00000144248
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr10_+_31357862 | 0.63 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr20_+_2625619 | 0.63 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr13_-_14798134 | 0.62 |
ENSDART00000020576
|
cdc25b
|
cell division cycle 25B |
| chr8_-_40149140 | 0.61 |
ENSDART00000005118
|
gpx8
|
glutathione peroxidase 8 (putative) |
| chr10_+_21487218 | 0.60 |
ENSDART00000147215
ENSDART00000019252 |
fbxw11b
|
F-box and WD repeat domain containing 11b |
| chr23_+_25953162 | 0.56 |
ENSDART00000138731
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr13_-_999570 | 0.54 |
ENSDART00000121970
|
pno1
|
partner of NOB1 homolog |
| chr8_+_53734037 | 0.53 |
ENSDART00000179055
|
ENSDARG00000106018
|
ENSDARG00000106018 |
| chr8_-_12394639 | 0.50 |
ENSDART00000133350
|
traf1
|
TNF receptor-associated factor 1 |
| chr11_-_13295436 | 0.50 |
|
|
|
| chr8_-_12394879 | 0.50 |
ENSDART00000101174
|
traf1
|
TNF receptor-associated factor 1 |
| chr23_-_27663219 | 0.50 |
ENSDART00000138381
|
si:ch211-156j22.4
|
si:ch211-156j22.4 |
| chr21_-_21604542 | 0.44 |
ENSDART00000010282
|
cbr1l
|
carbonyl reductase 1-like |
| chr13_+_6061419 | 0.44 |
|
|
|
| chr13_+_36831946 | 0.41 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr13_+_36832145 | 0.40 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr23_-_20592068 | 0.37 |
ENSDART00000153767
|
CR352228.1
|
ENSDARG00000097895 |
| chr9_-_34145854 | 0.36 |
|
|
|
| chr9_+_188598 | 0.36 |
ENSDART00000162764
|
rrp1
|
ribosomal RNA processing 1 |
| chr9_-_52033978 | 0.35 |
ENSDART00000148918
|
tank
|
TRAF family member-associated NFKB activator |
| chr5_-_31306608 | 0.34 |
|
|
|
| chr1_+_54059634 | 0.34 |
ENSDART00000162075
|
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr23_+_20592121 | 0.31 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr10_-_26263954 | 0.29 |
ENSDART00000079194
|
arfip2b
|
ADP-ribosylation factor interacting protein 2b |
| chr7_-_23928945 | 0.29 |
|
|
|
| chr16_-_13927947 | 0.29 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr1_+_48770878 | 0.28 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr16_-_13927684 | 0.27 |
ENSDART00000146719
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr21_-_41804961 | 0.26 |
ENSDART00000046076
|
ENSDARG00000030665
|
ENSDARG00000030665 |
| chr7_+_56171081 | 0.25 |
ENSDART00000073596
ENSDART00000123273 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr10_+_31358110 | 0.25 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr11_+_269859 | 0.24 |
ENSDART00000109091
|
mettl1
|
methyltransferase like 1 |
| chr5_+_23582396 | 0.22 |
ENSDART00000051549
ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr13_-_25689369 | 0.19 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr13_+_28545221 | 0.17 |
ENSDART00000027213
|
inaa
|
internexin neuronal intermediate filament protein, alpha a |
| chr9_+_32496059 | 0.16 |
ENSDART00000078513
|
mob4
|
MOB family member 4, phocein |
| chr5_+_29988157 | 0.12 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr2_+_25107108 | 0.11 |
ENSDART00000136072
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr9_-_28464146 | 0.09 |
ENSDART00000137582
|
creb1b
|
cAMP responsive element binding protein 1b |
| chr3_+_23587109 | 0.08 |
|
|
|
| chr20_-_35173500 | 0.08 |
ENSDART00000019196
|
lft1
|
lefty1 |
| chr9_+_20743306 | 0.05 |
ENSDART00000147435
|
man1a2
|
mannosidase, alpha, class 1A, member 2 |
| chr17_+_8018808 | 0.04 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr17_+_9918253 | 0.02 |
ENSDART00000169892
|
srp54
|
signal recognition particle 54 |
| chr25_-_21058792 | 0.01 |
ENSDART00000156257
|
wnk1a
|
WNK lysine deficient protein kinase 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.3 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.3 | 1.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.2 | 1.4 | GO:2000479 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.6 | GO:1902751 | regulation of mitotic cell cycle, embryonic(GO:0009794) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic cell cycle, embryonic(GO:0045448) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 3.7 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.2 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 1.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.2 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 4.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 2.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.7 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 4.7 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.8 | GO:0098802 | plasma membrane receptor complex(GO:0098802) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.3 | 1.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 4.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 2.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.8 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 0.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.7 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |