Project
DANIO-CODE
Navigation
Downloads
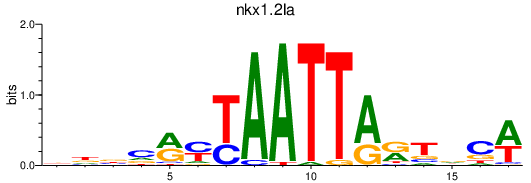
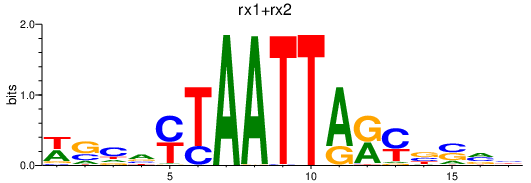
Results for nkx1.2la_rx1+rx2
Z-value: 0.26
Transcription factors associated with nkx1.2la_rx1+rx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx1.2la
|
ENSDARG00000006350 | NK1 transcription factor related 2-like,a |
|
rx2
|
ENSDARG00000040321 | retinal homeobox gene 2 |
|
rx1
|
ENSDARG00000071684 | retinal homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx2 | dr10_dc_chr2_-_55737505_55737528 | -0.13 | 6.3e-01 | Click! |
| nkx1.2la | dr10_dc_chr13_+_40644724_40644738 | -0.12 | 6.7e-01 | Click! |
| rx1 | dr10_dc_chr22_-_4622401_4622455 | -0.06 | 8.3e-01 | Click! |
Activity profile of nkx1.2la_rx1+rx2 motif
Sorted Z-values of nkx1.2la_rx1+rx2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx1.2la_rx1+rx2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_37303599 | 0.46 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr6_+_36862078 | 0.43 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr20_-_29961498 | 0.31 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr20_-_29961589 | 0.31 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr2_-_26941084 | 0.31 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_-_34058331 | 0.29 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr6_+_36861678 | 0.28 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr14_-_14353487 | 0.27 |
ENSDART00000172241
|
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr13_-_36672424 | 0.27 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr20_-_29961621 | 0.23 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr20_+_29306677 | 0.22 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr13_-_36672285 | 0.20 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr15_+_35089305 | 0.18 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr7_-_29906486 | 0.17 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr16_+_23884575 | 0.17 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr17_+_43041951 | 0.17 |
ENSDART00000154863
|
isca2
|
iron-sulfur cluster assembly 2 |
| chr18_-_40718244 | 0.16 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr5_+_37303707 | 0.15 |
ENSDART00000097754
ENSDART00000162470 |
tmprss4b
|
transmembrane protease, serine 4b |
| chr21_-_2350090 | 0.15 |
ENSDART00000168946
|
si:ch211-241b2.4
|
si:ch211-241b2.4 |
| chr7_-_73204361 | 0.15 |
ENSDART00000157500
|
AL929105.1
|
ENSDARG00000105194 |
| chr25_+_10450395 | 0.14 |
ENSDART00000067678
|
zgc:110339
|
zgc:110339 |
| chr24_-_14446593 | 0.14 |
|
|
|
| chr20_+_29306945 | 0.13 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr21_+_10757086 | 0.13 |
|
|
|
| chr1_+_35253862 | 0.12 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr17_-_36904594 | 0.12 |
|
|
|
| chr14_+_30455165 | 0.12 |
ENSDART00000144817
|
cfl1
|
cofilin 1 |
| chr16_-_55263338 | 0.11 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr2_-_26940965 | 0.10 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr24_-_24999240 | 0.10 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr20_-_37910887 | 0.10 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr15_+_34734212 | 0.10 |
ENSDART00000099776
|
tspan13a
|
tetraspanin 13a |
| chr20_+_29306863 | 0.09 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr11_-_38856849 | 0.09 |
ENSDART00000113185
|
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr19_+_5562075 | 0.09 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr23_-_19299279 | 0.09 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr7_-_29906449 | 0.09 |
ENSDART00000046689
|
tmed3
|
transmembrane p24 trafficking protein 3 |
| chr6_+_36862148 | 0.09 |
ENSDART00000104160
|
ENSDARG00000070746
|
ENSDARG00000070746 |
| chr8_+_3364453 | 0.09 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr14_+_46362661 | 0.08 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr2_+_25659945 | 0.08 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr18_-_43890836 | 0.08 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr13_-_18564182 | 0.08 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr1_+_51087450 | 0.08 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr19_+_21783111 | 0.08 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr7_-_40359613 | 0.07 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr8_+_11287550 | 0.07 |
ENSDART00000115057
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr11_-_2437396 | 0.07 |
|
|
|
| chr13_-_8360779 | 0.07 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr24_+_24999138 | 0.07 |
ENSDART00000152095
ENSDART00000154988 ENSDART00000156098 |
CR383672.2
|
ENSDARG00000096452 |
| chr1_+_14572155 | 0.07 |
ENSDART00000033018
|
pi4k2b
|
phosphatidylinositol 4-kinase type 2 beta |
| chr9_-_35127938 | 0.07 |
ENSDART00000077814
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr1_+_5464274 | 0.07 |
ENSDART00000144559
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr18_+_49974874 | 0.07 |
ENSDART00000126916
|
mob2b
|
MOB kinase activator 2b |
| chr10_-_4641624 | 0.06 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr13_-_36537892 | 0.06 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr17_+_16038358 | 0.06 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_-_19986607 | 0.06 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr20_+_36909452 | 0.06 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr25_-_1243081 | 0.06 |
ENSDART00000156062
|
calml4b
|
calmodulin-like 4b |
| chr23_+_20779487 | 0.06 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr23_-_33692244 | 0.05 |
|
|
|
| chr11_+_16017857 | 0.05 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr10_-_28230992 | 0.05 |
ENSDART00000134491
|
med13a
|
mediator complex subunit 13a |
| chr14_+_22159893 | 0.05 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr10_-_4641512 | 0.05 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr3_+_27667194 | 0.05 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr20_+_35176069 | 0.05 |
|
|
|
| chr13_-_36672606 | 0.05 |
ENSDART00000179242
|
sav1
|
salvador family WW domain containing protein 1 |
| chr18_-_5749619 | 0.05 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr5_+_12894381 | 0.05 |
ENSDART00000051669
|
tctn2
|
tectonic family member 2 |
| chr22_+_699477 | 0.05 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr24_+_16402587 | 0.05 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr19_+_1743359 | 0.05 |
ENSDART00000166744
|
dennd3a
|
DENN/MADD domain containing 3a |
| chr19_+_44185325 | 0.04 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr16_+_13928376 | 0.04 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr12_+_3632548 | 0.04 |
|
|
|
| chr19_+_21783204 | 0.04 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr6_+_40924559 | 0.04 |
ENSDART00000133599
|
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_+_50873843 | 0.04 |
|
|
|
| chr6_-_40924459 | 0.04 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr17_+_135261 | 0.04 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr10_-_2761480 | 0.04 |
ENSDART00000131749
|
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr8_-_17692185 | 0.04 |
|
|
|
| chr23_+_17220363 | 0.04 |
ENSDART00000143420
|
BX927275.2
|
ENSDARG00000095017 |
| chr16_-_25653129 | 0.03 |
ENSDART00000149411
|
atxn1b
|
ataxin 1b |
| chr6_+_344760 | 0.03 |
|
|
|
| chr10_+_40293111 | 0.03 |
|
|
|
| chr2_+_8981028 | 0.03 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr21_-_30045669 | 0.03 |
ENSDART00000155188
|
ccnjl
|
cyclin J-like |
| chr24_-_24999290 | 0.03 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr12_+_22459177 | 0.03 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr17_-_135071 | 0.03 |
|
|
|
| chr21_+_21158761 | 0.03 |
ENSDART00000058311
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr16_+_51597409 | 0.03 |
|
|
|
| chr8_-_12253026 | 0.03 |
ENSDART00000091612
|
dab2ipa
|
DAB2 interacting protein a |
| chr16_-_31393328 | 0.03 |
ENSDART00000178298
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr7_-_50137683 | 0.03 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr12_+_33257120 | 0.03 |
|
|
|
| chr13_+_4743399 | 0.03 |
ENSDART00000092521
|
micu1
|
mitochondrial calcium uptake 1 |
| chr20_+_29307283 | 0.02 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr8_-_53165501 | 0.02 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr5_-_25133456 | 0.02 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr3_+_23561306 | 0.02 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr22_-_19986461 | 0.02 |
ENSDART00000093310
|
celf5a
|
cugbp, Elav-like family member 5a |
| chr20_+_10502210 | 0.02 |
ENSDART00000155888
|
AL845550.1
|
ENSDARG00000097342 |
| chr12_-_33256671 | 0.02 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr15_-_5827067 | 0.02 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr4_+_5246465 | 0.02 |
ENSDART00000137966
|
ccdc167
|
coiled-coil domain containing 167 |
| chr14_+_5078937 | 0.02 |
ENSDART00000031508
|
lbx2
|
ladybird homeobox 2 |
| chr24_+_19270877 | 0.02 |
|
|
|
| chr3_-_23442774 | 0.02 |
|
|
|
| chr21_+_11742219 | 0.02 |
ENSDART00000081661
|
nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr9_-_32803650 | 0.01 |
|
|
|
| chr18_+_44635321 | 0.01 |
|
|
|
| chr24_+_37692365 | 0.01 |
|
|
|
| chr5_+_51992974 | 0.01 |
ENSDART00000170341
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr25_+_34433321 | 0.01 |
ENSDART00000042678
ENSDART00000158092 |
vps4a
|
vacuolar protein sorting 4a homolog A (S. cerevisiae) |
| chr19_+_32679320 | 0.01 |
|
|
|
| chr19_+_32679258 | 0.01 |
|
|
|
| chr2_+_54267946 | 0.01 |
ENSDART00000015535
|
ctnnbl1
|
catenin, beta like 1 |
| chr6_+_40925259 | 0.01 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr12_-_33256934 | 0.01 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_+_41910191 | 0.01 |
|
|
|
| chr7_-_57948889 | 0.01 |
|
|
|
| chr9_+_33177527 | 0.01 |
ENSDART00000174533
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr21_+_28441951 | 0.01 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr6_+_57544814 | 0.00 |
ENSDART00000157330
|
necab3
|
N-terminal EF-hand calcium binding protein 3 |
| chr20_-_16553010 | 0.00 |
|
|
|
| chr21_-_2316052 | 0.00 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr21_-_45323151 | 0.00 |
ENSDART00000151106
|
ENSDARG00000053404
|
ENSDARG00000053404 |
| chr2_-_2800076 | 0.00 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr1_+_53384116 | 0.00 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr20_+_28958586 | 0.00 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr24_-_14447136 | 0.00 |
|
|
|
| chr1_+_35253821 | 0.00 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr8_-_25015215 | 0.00 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.9 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:0071451 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.0 | 0.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |