Project
DANIO-CODE
Navigation
Downloads
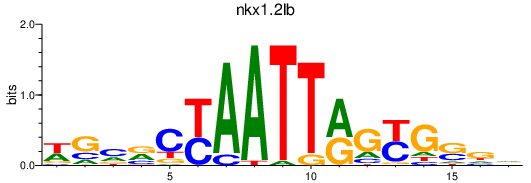
Results for nkx1.2lb
Z-value: 0.30
Transcription factors associated with nkx1.2lb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx1.2lb
|
ENSDARG00000099427 | NK1 transcription factor related 2-like,b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx1.2lb | dr10_dc_chr14_+_14759314_14759320 | -0.48 | 6.1e-02 | Click! |
Activity profile of nkx1.2lb motif
Sorted Z-values of nkx1.2lb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx1.2lb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34058331 | 1.29 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr8_-_23759076 | 1.06 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr20_-_14218080 | 0.93 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| KN150266v1_-_68652 | 0.75 |
|
|
|
| chr20_+_14218237 | 0.63 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| KN150663v1_-_3381 | 0.60 |
|
|
|
| chr13_-_18564182 | 0.52 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr2_-_26941084 | 0.51 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr2_+_6341404 | 0.45 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr13_-_36672424 | 0.39 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr20_-_14218236 | 0.35 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| KN150266v1_-_68620 | 0.35 |
|
|
|
| chr23_-_33692244 | 0.34 |
|
|
|
| chr18_-_43890836 | 0.34 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr8_-_3974320 | 0.33 |
ENSDART00000159895
|
mtmr3
|
myotubularin related protein 3 |
| chr16_+_25693878 | 0.32 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr12_-_4212149 | 0.30 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr13_+_12629211 | 0.29 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr6_+_31381325 | 0.27 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr5_-_32698092 | 0.26 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr14_+_46362661 | 0.26 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr7_+_20132424 | 0.25 |
ENSDART00000173809
|
si:dkey-33c9.6
|
si:dkey-33c9.6 |
| chr20_-_37910887 | 0.24 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr6_+_31381039 | 0.24 |
ENSDART00000038990
|
jak1
|
Janus kinase 1 |
| chr13_-_30011574 | 0.24 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr11_+_12754166 | 0.24 |
ENSDART00000123445
ENSDART00000163364 ENSDART00000066122 |
rtel1
|
regulator of telomere elongation helicase 1 |
| chr10_-_34969570 | 0.22 |
|
|
|
| chr12_-_4212095 | 0.22 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr14_-_30604897 | 0.22 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr13_-_36537892 | 0.21 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr11_+_16017857 | 0.20 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr21_+_10757086 | 0.19 |
|
|
|
| chr5_+_32698516 | 0.19 |
ENSDART00000097945
|
usp20
|
ubiquitin specific peptidase 20 |
| chr2_-_26940965 | 0.19 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr23_+_34120812 | 0.18 |
ENSDART00000143933
|
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr12_-_7201660 | 0.18 |
ENSDART00000152450
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr12_-_18997267 | 0.17 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr17_+_5454020 | 0.16 |
ENSDART00000097455
|
fam167aa
|
family with sequence similarity 167, member Aa |
| chr14_+_30073402 | 0.16 |
ENSDART00000136009
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr22_-_6972182 | 0.16 |
|
|
|
| chr13_-_6151100 | 0.16 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr16_+_32097505 | 0.16 |
ENSDART00000039109
|
leng1
|
leukocyte receptor cluster (LRC) member 1 |
| chr13_-_30011613 | 0.15 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr23_+_29431388 | 0.14 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr7_-_40359613 | 0.14 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr14_+_8288174 | 0.14 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| KN150703v1_+_15225 | 0.13 |
|
|
|
| chr2_+_10209233 | 0.13 |
ENSDART00000160304
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr13_-_36672285 | 0.12 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr2_+_6341345 | 0.12 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr17_-_36904594 | 0.12 |
|
|
|
| chr5_+_60774642 | 0.12 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr18_+_13267740 | 0.11 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr21_-_30977021 | 0.10 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr2_-_48203102 | 0.10 |
|
|
|
| chr13_-_36538099 | 0.09 |
|
|
|
| chr21_-_29995378 | 0.08 |
ENSDART00000178201
|
pwwp2a
|
PWWP domain containing 2A |
| chr13_+_12629243 | 0.07 |
ENSDART00000015127
|
zgc:100846
|
zgc:100846 |
| chr22_+_39110524 | 0.07 |
ENSDART00000153841
|
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr6_+_17928720 | 0.07 |
ENSDART00000124948
|
srp68
|
signal recognition particle 68 |
| chr14_+_46318771 | 0.06 |
ENSDART00000034606
ENSDART00000173301 |
sfxn5b
|
sideroflexin 5b |
| chr2_+_50873843 | 0.06 |
|
|
|
| chr6_+_3313515 | 0.06 |
ENSDART00000011785
|
rad54l
|
RAD54-like (S. cerevisiae) |
| chr7_-_73624022 | 0.05 |
ENSDART00000111049
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr3_+_52373536 | 0.05 |
|
|
|
| chr5_-_21548930 | 0.04 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr20_+_36909452 | 0.04 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr3_+_32700750 | 0.04 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr8_-_23759198 | 0.03 |
ENSDART00000145894
|
zgc:195245
|
zgc:195245 |
| chr13_-_36672606 | 0.03 |
ENSDART00000179242
|
sav1
|
salvador family WW domain containing protein 1 |
| chr3_+_32700882 | 0.03 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr5_-_13267041 | 0.02 |
|
|
|
| chr9_+_26218474 | 0.02 |
ENSDART00000177394
|
CR391949.1
|
ENSDARG00000107108 |
| chr16_+_10950751 | 0.01 |
ENSDART00000128762
|
dedd1
|
death effector domain-containing 1 |
| chr18_+_13267658 | 0.01 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr4_+_63623569 | 0.01 |
ENSDART00000170824
|
znf1108
|
zinc finger protein 1108 |
| chr4_-_47142181 | 0.00 |
ENSDART00000150426
|
znf1064
|
zinc finger protein 1064 |
| chr17_-_15181427 | 0.00 |
ENSDART00000002176
ENSDART00000138764 |
wdhd1
|
WD repeat and HMG-box DNA binding protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.1 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.5 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 0.4 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.6 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.1 | 0.5 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 0.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |