Project
DANIO-CODE
Navigation
Downloads
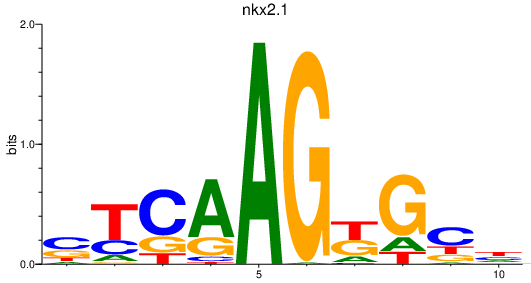
Results for nkx2.1
Z-value: 0.24
Transcription factors associated with nkx2.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.1
|
ENSDARG00000019835 | NK2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.1 | dr10_dc_chr17_+_38314814_38314876 | -0.50 | 4.9e-02 | Click! |
Activity profile of nkx2.1 motif
Sorted Z-values of nkx2.1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_22450582 | 0.65 |
|
|
|
| chr19_-_33264349 | 0.38 |
|
|
|
| chr20_+_9141086 | 0.35 |
ENSDART00000064144
ENSDART00000144396 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr6_+_38882608 | 0.31 |
ENSDART00000019939
|
bin2b
|
bridging integrator 2b |
| chr3_+_17783319 | 0.28 |
ENSDART00000104299
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr3_+_17783868 | 0.26 |
ENSDART00000165644
|
cnp
|
2',3'-cyclic nucleotide 3' phosphodiesterase |
| chr23_-_18105560 | 0.23 |
|
|
|
| chr14_+_10351071 | 0.20 |
ENSDART00000061932
|
atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr2_-_11335644 | 0.20 |
ENSDART00000135450
|
cryz
|
crystallin, zeta (quinone reductase) |
| chr15_-_16137131 | 0.20 |
|
|
|
| chr4_-_5786680 | 0.19 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr7_+_34235297 | 0.19 |
ENSDART00000173957
|
rfx7
|
regulatory factor X, 7 |
| chr20_-_53123015 | 0.19 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr17_-_49256667 | 0.18 |
ENSDART00000112806
|
orc3
|
origin recognition complex, subunit 3 |
| chr22_-_20899539 | 0.17 |
ENSDART00000100642
|
ell
|
elongation factor RNA polymerase II |
| chr3_+_22204475 | 0.17 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr11_+_25358353 | 0.16 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr21_-_2322963 | 0.16 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr14_+_15778201 | 0.14 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr23_+_37364185 | 0.14 |
ENSDART00000006436
|
pink1
|
PTEN induced putative kinase 1 |
| chr15_-_29079613 | 0.13 |
ENSDART00000060018
|
dharma
|
dharma |
| chr1_+_15928362 | 0.13 |
ENSDART00000149026
ENSDART00000159785 ENSDART00000164899 |
pcm1
|
pericentriolar material 1 |
| chr1_+_13333864 | 0.12 |
ENSDART00000079716
|
hpf1
|
histone PARylation factor 1 |
| chr3_-_30757570 | 0.12 |
ENSDART00000124458
|
suv420h2
|
suppressor of variegation 4-20 homolog 2 (Drosophila) |
| chr17_-_12095723 | 0.11 |
ENSDART00000115321
|
ahctf1
|
AT hook containing transcription factor 1 |
| chr6_+_38882803 | 0.11 |
ENSDART00000132798
|
bin2b
|
bridging integrator 2b |
| chr19_-_25842574 | 0.11 |
ENSDART00000036854
|
glcci1
|
glucocorticoid induced 1 |
| chr19_-_38832985 | 0.11 |
|
|
|
| chr22_+_15872131 | 0.11 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr9_-_30692223 | 0.10 |
|
|
|
| chr7_+_25554355 | 0.10 |
ENSDART00000149835
|
mtm1
|
myotubularin 1 |
| chr9_-_27581087 | 0.10 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr23_-_33753484 | 0.10 |
ENSDART00000138416
|
tfcp2
|
transcription factor CP2 |
| chr8_-_51380776 | 0.10 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr11_+_23807915 | 0.10 |
ENSDART00000006703
|
maf1
|
MAF1 homolog, negative regulator of RNA polymerase III |
| chr19_+_7373519 | 0.09 |
ENSDART00000026634
|
ccdc127b
|
coiled-coil domain containing 127b |
| chr11_+_25358389 | 0.09 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr8_+_45339021 | 0.09 |
|
|
|
| chr2_-_11335695 | 0.09 |
ENSDART00000135450
|
cryz
|
crystallin, zeta (quinone reductase) |
| chr3_+_22204569 | 0.09 |
ENSDART00000055676
|
ENSDARG00000038186
|
ENSDARG00000038186 |
| chr12_+_31379514 | 0.08 |
ENSDART00000153390
|
gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr6_-_22060441 | 0.08 |
ENSDART00000167167
|
utp6
|
UTP6, small subunit (SSU) processome component, homolog (yeast) |
| chr7_-_24778185 | 0.07 |
ENSDART00000135179
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr3_-_23331479 | 0.07 |
ENSDART00000040065
|
casc3
|
cancer susceptibility candidate 3 |
| chr25_-_20593345 | 0.07 |
ENSDART00000158130
|
edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae) |
| chr2_-_37419017 | 0.07 |
ENSDART00000015723
|
prkci
|
protein kinase C, iota |
| chr5_-_34856057 | 0.06 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr18_-_7488960 | 0.05 |
ENSDART00000052803
|
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr20_+_31375056 | 0.04 |
|
|
|
| chr1_-_22603959 | 0.04 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr18_-_321265 | 0.04 |
ENSDART00000136693
|
tm2d3
|
TM2 domain containing 3 |
| chr22_-_20695416 | 0.04 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr1_+_50348973 | 0.04 |
|
|
|
| chr24_-_8637129 | 0.04 |
ENSDART00000082362
|
mak
|
male germ cell-associated kinase |
| chr11_-_35778578 | 0.02 |
ENSDART00000112684
|
setmar
|
SET domain and mariner transposase fusion gene |
| chr12_+_34851952 | 0.02 |
ENSDART00000123988
|
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr7_+_48402923 | 0.02 |
ENSDART00000174084
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr18_-_895821 | 0.02 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr18_+_29166912 | 0.01 |
ENSDART00000089031
|
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr7_-_15781823 | 0.01 |
ENSDART00000002498
|
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr3_-_23331363 | 0.01 |
ENSDART00000040065
|
casc3
|
cancer susceptibility candidate 3 |
| chr7_-_28300008 | 0.01 |
|
|
|
| chr2_-_22692538 | 0.00 |
|
|
|
| chr6_-_6091628 | 0.00 |
ENSDART00000132350
|
rtn4a
|
reticulon 4a |
| chr13_+_46637827 | 0.00 |
ENSDART00000167931
|
mtrf1l
|
mitochondrial translational release factor 1-like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0051292 | pore complex assembly(GO:0046931) nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.3 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 0.0 | 0.5 | GO:0004112 | cyclic-nucleotide phosphodiesterase activity(GO:0004112) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.0 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.0 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |