Project
DANIO-CODE
Navigation
Downloads
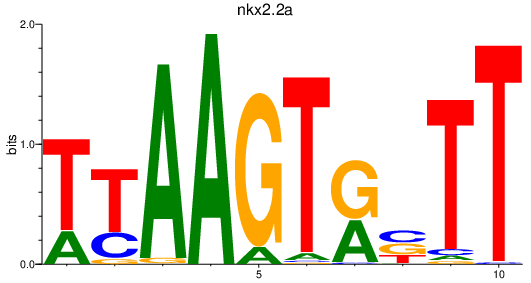
Results for nkx2.2a
Z-value: 0.80
Transcription factors associated with nkx2.2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2a
|
ENSDARG00000053298 | NK2 homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2a | dr10_dc_chr17_-_42778156_42778239 | 0.84 | 4.6e-05 | Click! |
Activity profile of nkx2.2a motif
Sorted Z-values of nkx2.2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_69399273 | 2.14 |
ENSDART00000159823
ENSDART00000075178 ENSDART00000168942 ENSDART00000126739 |
tspan5a
|
tetraspanin 5a |
| chr1_+_51925783 | 1.87 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr8_-_9081109 | 1.74 |
ENSDART00000176850
|
slc6a8
|
solute carrier family 6 (neurotransmitter transporter), member 8 |
| chr14_-_26238386 | 1.63 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr20_-_54206610 | 1.55 |
ENSDART00000004756
|
hsp90aa1.1
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 1 |
| chr8_+_23464087 | 1.54 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr5_+_37053530 | 1.41 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr23_+_22763861 | 1.38 |
|
|
|
| chr6_+_4712785 | 1.37 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr10_-_10059864 | 1.35 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr9_-_22347397 | 1.33 |
ENSDART00000146188
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr10_+_4094568 | 1.31 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr2_+_49358871 | 1.31 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr13_-_18506153 | 1.29 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr15_-_29454583 | 1.29 |
ENSDART00000145976
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr19_+_38834972 | 1.27 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr10_+_21850059 | 1.24 |
ENSDART00000164634
ENSDART00000172513 |
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr4_-_24152926 | 1.21 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr19_+_1626598 | 1.21 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr16_+_737615 | 1.21 |
ENSDART00000161774
|
irx1a
|
iroquois homeobox 1a |
| chr8_+_29953399 | 1.19 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr6_-_10593395 | 1.19 |
ENSDART00000020261
|
chrna1
|
cholinergic receptor, nicotinic, alpha 1 (muscle) |
| chr9_-_11589126 | 1.18 |
ENSDART00000146832
|
cryba2b
|
crystallin, beta A2b |
| chr16_+_23172295 | 1.18 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr23_+_19887319 | 1.16 |
ENSDART00000139192
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr16_+_21109486 | 1.14 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr21_-_40694583 | 1.13 |
ENSDART00000045124
|
pomp
|
proteasome maturation protein |
| chr19_+_38834765 | 1.08 |
ENSDART00000140198
ENSDART00000033037 ENSDART00000035093 |
col9a2
|
procollagen, type IX, alpha 2 |
| chr9_+_21911860 | 1.07 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr2_-_24614261 | 1.06 |
ENSDART00000099532
|
vmhc
|
ventricular myosin heavy chain |
| chr1_+_14451504 | 1.05 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr5_-_58269742 | 1.04 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr6_+_18588827 | 1.03 |
|
|
|
| chr16_+_14817799 | 1.01 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr6_+_40631517 | 1.00 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr23_+_6861489 | 0.99 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr6_+_3771586 | 0.98 |
|
|
|
| chr25_+_30746445 | 0.97 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr15_-_4537178 | 0.97 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr5_+_29578638 | 0.95 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| chr6_+_19167652 | 0.94 |
ENSDART00000086619
|
prkca
|
protein kinase C, alpha |
| chr8_-_2447758 | 0.93 |
ENSDART00000101125
|
rpl6
|
ribosomal protein L6 |
| chr23_+_25428372 | 0.92 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr13_-_14978365 | 0.92 |
|
|
|
| chr12_-_8780098 | 0.92 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr23_+_26995808 | 0.92 |
|
|
|
| chr14_+_4810147 | 0.92 |
ENSDART00000114678
|
nanos1
|
nanos homolog 1 |
| chr9_+_6684835 | 0.91 |
|
|
|
| chr22_+_20695983 | 0.90 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr2_-_55737505 | 0.90 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr9_+_24198112 | 0.88 |
ENSDART00000148226
|
mlphb
|
melanophilin b |
| chr25_+_3232912 | 0.88 |
ENSDART00000104877
ENSDART00000170765 ENSDART00000158571 |
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr14_-_425659 | 0.87 |
|
|
|
| chr23_-_31585883 | 0.87 |
ENSDART00000157511
|
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr7_-_37816600 | 0.86 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr7_-_32604392 | 0.86 |
ENSDART00000132504
|
ano5b
|
anoctamin 5b |
| chr20_-_25652576 | 0.84 |
ENSDART00000003666
ENSDART00000122351 ENSDART00000063081 ENSDART00000153282 |
cyp2ad3
|
cytochrome P450, family 2, subfamily AD, polypeptide 3 |
| chr24_+_5906087 | 0.84 |
ENSDART00000131768
|
abi1a
|
abl-interactor 1a |
| chr7_-_29300402 | 0.82 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr11_+_42821961 | 0.81 |
ENSDART00000013642
|
foxg1b
|
forkhead box G1b |
| chr5_-_47106694 | 0.81 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr16_+_19926776 | 0.80 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr22_+_38096040 | 0.80 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr3_-_27516974 | 0.79 |
ENSDART00000151675
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr24_+_3931981 | 0.78 |
ENSDART00000167043
|
pfkpa
|
phosphofructokinase, platelet a |
| chr23_-_7892862 | 0.78 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr16_+_14817718 | 0.77 |
ENSDART00000027982
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr7_-_29300448 | 0.77 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr3_-_58161663 | 0.77 |
ENSDART00000178787
|
CABZ01038493.1
|
ENSDARG00000108952 |
| chr17_+_7438152 | 0.77 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr16_-_41994015 | 0.77 |
|
|
|
| chr15_-_12484651 | 0.76 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr7_+_19857885 | 0.76 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr24_-_35897257 | 0.76 |
|
|
|
| KN150683v1_+_13359 | 0.74 |
|
|
|
| chr3_-_15849644 | 0.74 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr1_+_25662910 | 0.73 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr12_-_10001043 | 0.73 |
ENSDART00000152250
|
ngfrb
|
nerve growth factor receptor b |
| chr25_+_18467835 | 0.73 |
ENSDART00000172338
|
cav1
|
caveolin 1 |
| chr13_-_29294141 | 0.73 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr14_+_21563827 | 0.73 |
ENSDART00000032141
|
spon2a
|
spondin 2a, extracellular matrix protein |
| chr22_+_28496424 | 0.72 |
ENSDART00000089546
|
abi3bpb
|
ABI family, member 3 (NESH) binding protein b |
| chr13_+_17333810 | 0.71 |
ENSDART00000134181
|
CT025897.1
|
ENSDARG00000093577 |
| chr4_+_2215426 | 0.70 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr4_-_12915292 | 0.70 |
ENSDART00000067135
|
msrb3
|
methionine sulfoxide reductase B3 |
| chr3_+_48362748 | 0.69 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr3_+_12432686 | 0.69 |
|
|
|
| chr12_+_48761772 | 0.68 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr22_+_21221777 | 0.68 |
ENSDART00000079046
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr12_-_9256949 | 0.67 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr23_-_31419805 | 0.67 |
ENSDART00000138106
|
phip
|
pleckstrin homology domain interacting protein |
| chr5_-_54954988 | 0.67 |
ENSDART00000142912
|
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr11_+_25266623 | 0.67 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr7_+_19857748 | 0.66 |
ENSDART00000146335
|
zgc:114045
|
zgc:114045 |
| chr25_-_16658906 | 0.66 |
ENSDART00000124729
ENSDART00000110859 |
ribc2
|
RIB43A domain with coiled-coils 2 |
| chr9_+_24089828 | 0.66 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr8_-_25055160 | 0.65 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr17_-_20124053 | 0.65 |
ENSDART00000133327
|
actn2b
|
actinin, alpha 2b |
| chr12_-_27032151 | 0.65 |
ENSDART00000153365
|
BX001014.2
|
ENSDARG00000096750 |
| chr6_-_47507750 | 0.64 |
ENSDART00000128700
|
im:7151449
|
im:7151449 |
| chr8_-_25055333 | 0.64 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr18_-_8697347 | 0.64 |
ENSDART00000093131
|
FRMD4A
|
FERM domain containing 4A |
| chr23_+_24778062 | 0.64 |
|
|
|
| chr3_+_45375639 | 0.63 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr1_+_21309690 | 0.63 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr17_+_25659010 | 0.63 |
ENSDART00000029703
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr21_-_44610563 | 0.63 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr4_-_25847471 | 0.62 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr9_-_46272322 | 0.62 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr18_-_14723138 | 0.61 |
ENSDART00000010129
|
pdf
|
peptide deformylase (mitochondrial) |
| chr6_+_49096966 | 0.61 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr3_-_12432661 | 0.60 |
|
|
|
| chr13_-_40600924 | 0.60 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr12_+_30053414 | 0.60 |
ENSDART00000138252
|
ablim1b
|
actin binding LIM protein 1b |
| chr21_-_41445163 | 0.59 |
ENSDART00000170457
|
hmp19
|
HMP19 protein |
| chr8_+_51061523 | 0.58 |
ENSDART00000166249
|
DISP3
|
dispatched RND transporter family member 3 |
| chr7_-_28278286 | 0.56 |
ENSDART00000113313
|
st5
|
suppression of tumorigenicity 5 |
| chr25_+_35602451 | 0.56 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr9_-_23346038 | 0.55 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr2_+_2374651 | 0.55 |
ENSDART00000132500
|
c8a
|
complement component 8, alpha polypeptide |
| chr16_+_10531926 | 0.55 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr20_+_14591802 | 0.54 |
|
|
|
| chr12_-_45714172 | 0.54 |
ENSDART00000149044
|
pax2b
|
paired box 2b |
| chr22_-_17026124 | 0.53 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr10_-_39210658 | 0.53 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr23_+_22763789 | 0.53 |
|
|
|
| chr23_+_4423579 | 0.53 |
ENSDART00000039829
|
zgc:112175
|
zgc:112175 |
| chr7_+_20215723 | 0.52 |
ENSDART00000173724
ENSDART00000173773 |
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr14_-_25688444 | 0.52 |
ENSDART00000172909
|
atox1
|
antioxidant 1 copper chaperone |
| chr19_+_21140010 | 0.52 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr2_+_20814144 | 0.52 |
ENSDART00000177709
|
agla
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase a |
| chr7_-_41209891 | 0.52 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr4_-_75675696 | 0.52 |
ENSDART00000111941
|
zgc:174310
|
zgc:174310 |
| chr15_+_6789662 | 0.51 |
ENSDART00000013278
|
sirt2
|
sirtuin 2 (silent mating type information regulation 2, homolog) 2 (S. cerevisiae) |
| chr22_-_15576473 | 0.51 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr21_+_29190291 | 0.51 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr21_-_36338704 | 0.50 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr7_-_38427334 | 0.50 |
ENSDART00000173900
|
cd59
|
CD59 molecule (CD59 blood group) |
| chr8_+_20592039 | 0.50 |
ENSDART00000160421
|
nfic
|
nuclear factor I/C |
| chr12_+_33261056 | 0.50 |
ENSDART00000124982
|
ccdc57
|
coiled-coil domain containing 57 |
| chr2_+_22875548 | 0.49 |
ENSDART00000157884
|
hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr20_+_27121126 | 0.49 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr8_-_45752084 | 0.49 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr13_-_49695322 | 0.49 |
|
|
|
| chr2_-_11879076 | 0.49 |
ENSDART00000145108
|
ENSDARG00000039203
|
ENSDARG00000039203 |
| chr22_-_17026421 | 0.48 |
ENSDART00000138382
|
nfia
|
nuclear factor I/A |
| chr23_-_24468191 | 0.48 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr6_-_9933233 | 0.48 |
ENSDART00000151016
|
b3galt1a
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1a |
| chr4_-_7803189 | 0.48 |
ENSDART00000103070
|
cdk17
|
cyclin-dependent kinase 17 |
| chr8_-_14446883 | 0.47 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr11_-_16067646 | 0.47 |
|
|
|
| chr13_+_44720106 | 0.47 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr9_+_22953351 | 0.47 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr23_-_36812319 | 0.47 |
ENSDART00000109976
|
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr24_+_16924647 | 0.47 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr3_-_15850290 | 0.45 |
ENSDART00000160628
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr20_+_53310252 | 0.45 |
ENSDART00000175125
|
CABZ01089116.1
|
ENSDARG00000106770 |
| chr13_-_24248675 | 0.45 |
ENSDART00000046360
|
rhoua
|
ras homolog family member Ua |
| chr22_-_17561082 | 0.45 |
ENSDART00000060786
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr23_+_31420174 | 0.45 |
|
|
|
| chr11_-_11534819 | 0.45 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr16_+_34574302 | 0.45 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr1_+_23664646 | 0.45 |
ENSDART00000143428
|
rps3a
|
ribosomal protein S3A |
| chr9_-_41982635 | 0.44 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr16_+_11138924 | 0.44 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr5_+_35198499 | 0.44 |
|
|
|
| chr5_-_63521649 | 0.44 |
ENSDART00000029364
|
ak5l
|
adenylate kinase 5, like |
| chr19_+_24455545 | 0.44 |
ENSDART00000144541
|
rit1
|
Ras-like without CAAX 1 |
| chr4_-_16461748 | 0.43 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr16_+_11138879 | 0.43 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr7_-_33678058 | 0.42 |
ENSDART00000173513
|
pias1a
|
protein inhibitor of activated STAT, 1a |
| chr10_+_36710393 | 0.42 |
|
|
|
| chr10_-_8029671 | 0.42 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr3_+_24003840 | 0.42 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr13_-_22641001 | 0.42 |
|
|
|
| chr12_-_36415239 | 0.41 |
|
|
|
| chr14_+_23917724 | 0.41 |
ENSDART00000138082
ENSDART00000079215 |
stc2a
|
stanniocalcin 2a |
| chr11_-_9479592 | 0.41 |
ENSDART00000175829
|
CU179643.1
|
ENSDARG00000086489 |
| chr21_-_27164751 | 0.41 |
ENSDART00000065402
|
zgc:77262
|
zgc:77262 |
| chr11_-_3846064 | 0.41 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr11_-_25181234 | 0.40 |
ENSDART00000013714
|
gata1a
|
GATA binding protein 1a |
| chr24_-_21844170 | 0.40 |
ENSDART00000032963
|
apoob
|
apolipoprotein O, b |
| chr14_+_20614087 | 0.40 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr15_+_28173909 | 0.40 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr5_-_44243476 | 0.39 |
ENSDART00000161408
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr3_-_32795866 | 0.39 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr2_-_56321500 | 0.39 |
|
|
|
| chr14_-_45887648 | 0.39 |
ENSDART00000074208
|
cltb
|
clathrin, light chain B |
| chr6_-_41087828 | 0.39 |
ENSDART00000028217
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr18_+_20236794 | 0.38 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr23_+_24778090 | 0.38 |
|
|
|
| chr7_-_26246473 | 0.37 |
ENSDART00000140694
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr14_+_36529003 | 0.37 |
ENSDART00000111674
|
F2RL3
|
F2R like thrombin or trypsin receptor 3 |
| chr9_-_33972623 | 0.37 |
ENSDART00000008737
|
efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chr9_-_18903879 | 0.37 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr10_-_39211281 | 0.37 |
ENSDART00000148825
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr3_-_7220340 | 0.37 |
|
|
|
| chr14_+_16632329 | 0.37 |
ENSDART00000163013
|
limch1b
|
LIM and calponin homology domains 1b |
| chr10_-_5135388 | 0.36 |
ENSDART00000108587
|
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr4_+_71392976 | 0.36 |
ENSDART00000167082
|
zgc:152938
|
zgc:152938 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.3 | 0.9 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) regulation of nonmotile primary cilium assembly(GO:1902855) |
| 0.3 | 1.7 | GO:1903428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.2 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.2 | 1.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 1.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.2 | 1.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.6 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.2 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.7 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 0.5 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.2 | 0.5 | GO:2000378 | regulation of DNA binding(GO:0051101) regulation of oligodendrocyte progenitor proliferation(GO:0070445) negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.2 | 1.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.2 | 0.5 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 1.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.2 | 1.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.2 | 0.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.5 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.7 | GO:0019857 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.1 | 0.9 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.4 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.6 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.5 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.1 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) positive regulation of behavior(GO:0048520) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.8 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0030643 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.6 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.9 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 1.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.6 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.1 | 0.4 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.5 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.6 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 0.4 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.1 | 0.3 | GO:0061469 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 2.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 3.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.9 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.5 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.5 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.4 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 1.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.5 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 5.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 2.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.9 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.2 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.4 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.5 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 3.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.6 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.4 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.8 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.7 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 2.1 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.4 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.3 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.3 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.5 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 2.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.4 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 1.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 6.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 12.0 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0022626 | cytosolic ribosome(GO:0022626) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.9 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.2 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.2 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.2 | 1.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 0.6 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.2 | 0.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 0.8 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 0.5 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.5 | GO:0004937 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.8 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.5 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.1 | 0.4 | GO:0032028 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.5 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 2.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.1 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 2.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.1 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 4.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0030580 | 2-polyprenyl-6-methoxy-1,4-benzoquinone methyltransferase activity(GO:0008425) quinone cofactor methyltransferase activity(GO:0030580) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.6 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 0.8 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 0.8 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 2.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |