Project
DANIO-CODE
Navigation
Downloads
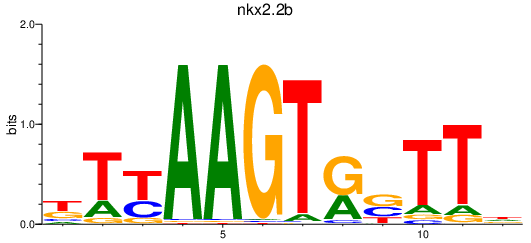
Results for nkx2.2b
Z-value: 1.03
Transcription factors associated with nkx2.2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.2b
|
ENSDARG00000101549 | NK2 homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.2b | dr10_dc_chr20_+_48950833_48950877 | -0.37 | 1.6e-01 | Click! |
Activity profile of nkx2.2b motif
Sorted Z-values of nkx2.2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_24971633 | 2.08 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr23_-_6831711 | 1.95 |
ENSDART00000125393
|
FP102169.1
|
ENSDARG00000089210 |
| chr1_+_15875513 | 1.94 |
|
|
|
| chr1_-_46141359 | 1.67 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr2_-_53797472 | 1.66 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr12_+_29121368 | 1.58 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr14_-_10311891 | 1.56 |
ENSDART00000136649
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr12_-_22279843 | 1.44 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr14_+_23420053 | 1.36 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr4_+_828545 | 1.34 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr13_+_33578737 | 1.29 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr5_-_30114979 | 1.21 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr23_-_24468191 | 1.13 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr23_-_24468290 | 1.10 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr8_+_26273354 | 1.10 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr2_-_53797503 | 1.04 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr3_-_27516974 | 1.04 |
ENSDART00000151675
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr18_-_35432838 | 0.96 |
ENSDART00000141703
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr9_-_18903879 | 0.96 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr7_+_22386456 | 0.88 |
ENSDART00000146081
|
ponzr5
|
plac8 onzin related protein 5 |
| chr25_-_14472107 | 0.88 |
|
|
|
| chr3_+_28450576 | 0.87 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr15_-_5912236 | 0.83 |
|
|
|
| chr22_+_866803 | 0.82 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr13_+_8509344 | 0.79 |
ENSDART00000140489
|
epcam
|
epithelial cell adhesion molecule |
| chr12_-_10438255 | 0.74 |
ENSDART00000106172
|
rac1a
|
ras-related C3 botulinum toxin substrate 1a (rho family, small GTP binding protein Rac1) |
| chr12_+_30119447 | 0.68 |
ENSDART00000102081
|
afap1l2
|
actin filament associated protein 1-like 2 |
| chr21_+_130546 | 0.68 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr10_+_33758581 | 0.68 |
ENSDART00000141650
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr12_-_28680035 | 0.67 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr9_-_20743808 | 0.66 |
|
|
|
| chr5_-_30115110 | 0.64 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr19_-_5142209 | 0.64 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr7_-_22519196 | 0.64 |
ENSDART00000129919
|
ENSDARG00000091111
|
ENSDARG00000091111 |
| chr11_-_25019298 | 0.64 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr16_-_2642235 | 0.64 |
|
|
|
| chr2_+_37441802 | 0.63 |
ENSDART00000132427
|
phc3
|
polyhomeotic homolog 3 (Drosophila) |
| chr24_+_9898743 | 0.63 |
ENSDART00000144186
|
pou6f2
|
POU class 6 homeobox 2 |
| chr12_-_9400272 | 0.63 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr13_+_45387478 | 0.63 |
ENSDART00000019113
|
tmem57b
|
transmembrane protein 57b |
| chr18_-_3430278 | 0.63 |
ENSDART00000157669
|
capn5a
|
calpain 5a |
| chr6_-_45867149 | 0.61 |
ENSDART00000062459
|
rbm19
|
RNA binding motif protein 19 |
| chr2_+_49343278 | 0.61 |
ENSDART00000175147
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr23_+_36608246 | 0.60 |
ENSDART00000113179
|
tspan31
|
tetraspanin 31 |
| chr5_-_24013767 | 0.59 |
ENSDART00000003957
|
tia1l
|
cytotoxic granule-associated RNA binding protein 1, like |
| chr18_+_17503828 | 0.59 |
ENSDART00000161827
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr19_-_5141645 | 0.58 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr14_+_24543399 | 0.58 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr13_+_8508786 | 0.58 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr11_+_24919694 | 0.58 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr7_+_22386328 | 0.58 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr15_-_5912917 | 0.57 |
|
|
|
| chr6_-_26904787 | 0.57 |
ENSDART00000134259
|
hdlbpa
|
high density lipoprotein binding protein a |
| chr8_-_38284904 | 0.56 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr19_-_5142009 | 0.55 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr5_+_60028622 | 0.55 |
|
|
|
| chr7_+_26457055 | 0.55 |
|
|
|
| chr12_-_4264663 | 0.54 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| chr8_-_38284748 | 0.54 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr4_+_6024067 | 0.53 |
|
|
|
| chr3_-_26830876 | 0.53 |
|
|
|
| chr12_-_28679813 | 0.53 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr1_+_29919378 | 0.52 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr16_+_39292206 | 0.52 |
ENSDART00000127319
ENSDART00000102510 |
zgc:77056
|
zgc:77056 |
| chr21_+_130849 | 0.52 |
|
|
|
| chr7_-_71343686 | 0.51 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr16_-_4780901 | 0.51 |
ENSDART00000029485
|
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr8_+_47737851 | 0.50 |
ENSDART00000160166
|
si:ch211-243p7.3
|
si:ch211-243p7.3 |
| chr12_-_22279890 | 0.50 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr18_-_27334059 | 0.49 |
ENSDART00000028294
|
zgc:56106
|
zgc:56106 |
| chr10_+_45138195 | 0.48 |
ENSDART00000162566
|
h2afvb
|
H2A histone family, member Vb |
| chr25_-_4294947 | 0.48 |
|
|
|
| chr4_+_46379457 | 0.48 |
ENSDART00000162948
|
BX324155.1
|
ENSDARG00000104638 |
| chr4_+_2215426 | 0.48 |
ENSDART00000168370
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr7_-_17129072 | 0.48 |
|
|
|
| chr19_-_5142238 | 0.48 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr22_-_22234380 | 0.47 |
ENSDART00000020937
|
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr5_-_18941045 | 0.47 |
ENSDART00000141268
|
kctd10
|
potassium channel tetramerization domain containing 10 |
| chr5_-_12086593 | 0.46 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr22_+_11745592 | 0.46 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr3_+_60299938 | 0.46 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr5_-_66820811 | 0.46 |
ENSDART00000128050
|
ENSDARG00000042520
|
ENSDARG00000042520 |
| chr18_-_22745979 | 0.46 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr14_+_23419864 | 0.46 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr3_+_24004141 | 0.45 |
ENSDART00000155216
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr22_-_20695416 | 0.45 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr2_-_53797678 | 0.45 |
ENSDART00000057053
|
im:7138239
|
im:7138239 |
| chr20_+_52021000 | 0.45 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr24_-_32607801 | 0.45 |
ENSDART00000143781
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr18_-_7522421 | 0.45 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr24_+_14516577 | 0.44 |
|
|
|
| chr1_-_358810 | 0.44 |
ENSDART00000098627
|
pros1
|
protein S (alpha) |
| chr22_-_22234261 | 0.43 |
ENSDART00000113824
ENSDART00000169594 ENSDART00000158261 |
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr6_-_22869821 | 0.43 |
ENSDART00000167674
|
mif4gda
|
MIF4G domain containing a |
| chr4_-_26046562 | 0.43 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr20_+_53563189 | 0.43 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr14_-_23811519 | 0.42 |
ENSDART00000124255
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr8_+_52456064 | 0.42 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr18_-_14723138 | 0.42 |
ENSDART00000010129
|
pdf
|
peptide deformylase (mitochondrial) |
| chr3_-_9755642 | 0.41 |
ENSDART00000168366
|
crebbpb
|
CREB binding protein b |
| chr24_+_35297193 | 0.41 |
ENSDART00000075142
ENSDART00000158801 |
pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr22_-_25015224 | 0.41 |
ENSDART00000015512
|
polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr20_-_40357302 | 0.41 |
|
|
|
| chr4_-_20119812 | 0.41 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr18_-_22745846 | 0.41 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr3_+_19490148 | 0.40 |
|
|
|
| chr23_+_38314255 | 0.40 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr12_-_22279713 | 0.40 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr7_-_37816600 | 0.40 |
ENSDART00000052366
|
cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr15_+_25516764 | 0.40 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr7_-_26246798 | 0.40 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr16_-_21277161 | 0.40 |
|
|
|
| chr16_-_41718971 | 0.40 |
|
|
|
| chr8_-_25055333 | 0.39 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr5_+_60028657 | 0.39 |
|
|
|
| chr16_-_25485248 | 0.38 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr6_+_28215039 | 0.38 |
ENSDART00000104394
|
smx5
|
smx5 |
| chr5_+_40543353 | 0.37 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr8_-_24269848 | 0.37 |
|
|
|
| chr8_-_49739908 | 0.37 |
ENSDART00000138810
|
gkap1
|
G kinase anchoring protein 1 |
| chr8_-_38284959 | 0.37 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr10_-_25383262 | 0.36 |
ENSDART00000166348
|
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr20_-_40357240 | 0.36 |
|
|
|
| chr12_-_25058837 | 0.36 |
ENSDART00000135368
|
rhoq
|
ras homolog family member Q |
| chr23_+_38313746 | 0.35 |
ENSDART00000129593
|
znf217
|
zinc finger protein 217 |
| chr6_+_12849673 | 0.35 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr8_-_38284660 | 0.34 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_+_866633 | 0.33 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr5_-_12086676 | 0.33 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr8_-_38285034 | 0.33 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_13654658 | 0.32 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr23_-_12305794 | 0.32 |
|
|
|
| chr11_+_3240975 | 0.32 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr24_-_34794538 | 0.32 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr11_-_25019899 | 0.32 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr22_+_35156074 | 0.32 |
ENSDART00000130581
|
rnf13
|
ring finger protein 13 |
| chr20_+_53563389 | 0.32 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr16_-_4781028 | 0.31 |
ENSDART00000029485
|
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr8_+_3372903 | 0.31 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr7_-_5252575 | 0.31 |
|
|
|
| chr16_-_21277108 | 0.31 |
|
|
|
| chr5_+_40543392 | 0.30 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr3_-_19913032 | 0.30 |
|
|
|
| chr16_-_33151404 | 0.30 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr14_-_10065562 | 0.30 |
ENSDART00000003879
|
fam199x
|
family with sequence similarity 199, X-linked |
| chr14_+_23419894 | 0.30 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr16_-_11908084 | 0.29 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr5_+_69178270 | 0.29 |
ENSDART00000097359
|
dnajc25
|
DnaJ (Hsp40) homolog, subfamily C , member 25 |
| chr13_+_36469737 | 0.29 |
ENSDART00000164545
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr19_-_33264349 | 0.29 |
|
|
|
| chr15_+_24628191 | 0.29 |
ENSDART00000140658
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr3_+_18657831 | 0.29 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr16_-_21277206 | 0.28 |
|
|
|
| chr8_-_25055160 | 0.28 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr2_-_22875246 | 0.28 |
ENSDART00000159641
|
znf644a
|
zinc finger protein 644a |
| chr6_-_39220951 | 0.28 |
ENSDART00000133305
ENSDART00000166290 |
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr22_-_7412566 | 0.28 |
ENSDART00000161615
ENSDART00000158210 |
BX511034.5
|
ENSDARG00000099295 |
| chr4_+_74885660 | 0.28 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr7_-_71343519 | 0.28 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr13_-_22568419 | 0.28 |
ENSDART00000165617
|
glud1a
|
glutamate dehydrogenase 1a |
| chr24_+_41448854 | 0.28 |
|
|
|
| chr21_-_36934378 | 0.27 |
ENSDART00000065208
|
nop16
|
NOP16 nucleolar protein homolog (yeast) |
| chr3_+_4058776 | 0.27 |
|
|
|
| chr2_-_15656155 | 0.27 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr14_-_38532734 | 0.27 |
ENSDART00000133442
|
gsr
|
glutathione reductase |
| chr22_+_21292537 | 0.27 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr9_-_14302452 | 0.27 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr5_-_56533970 | 0.27 |
ENSDART00000171252
|
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr12_-_22387786 | 0.26 |
ENSDART00000109707
|
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr20_-_34742647 | 0.26 |
|
|
|
| chr7_-_11436543 | 0.26 |
|
|
|
| KN150423v1_-_9882 | 0.26 |
|
|
|
| chr5_-_55243980 | 0.26 |
ENSDART00000014049
|
wdr36
|
WD repeat domain 36 |
| chr13_-_33186976 | 0.25 |
ENSDART00000136701
|
tmem234
|
transmembrane protein 234 |
| chr20_+_32853879 | 0.25 |
ENSDART00000023006
|
fam84a
|
family with sequence similarity 84, member A |
| chr17_+_10592507 | 0.25 |
ENSDART00000097274
|
ATG14
|
autophagy related 14 |
| chr6_-_57542175 | 0.25 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr19_-_21192796 | 0.25 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr19_-_1977399 | 0.25 |
|
|
|
| chr9_-_10026818 | 0.25 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr3_+_19536018 | 0.25 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr19_-_35380449 | 0.24 |
|
|
|
| chr7_-_11436576 | 0.24 |
|
|
|
| chr6_-_49548896 | 0.24 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr4_+_67023074 | 0.24 |
ENSDART00000158749
|
zgc:173702
|
zgc:173702 |
| chr19_+_31280451 | 0.24 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr12_+_23691397 | 0.24 |
ENSDART00000066331
|
svila
|
supervillin a |
| chr23_+_1069029 | 0.24 |
ENSDART00000159263
|
slc34a2b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2b |
| chr11_+_24919823 | 0.24 |
ENSDART00000162618
ENSDART00000128126 |
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr1_-_46430811 | 0.23 |
ENSDART00000048445
|
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr6_-_12967003 | 0.23 |
|
|
|
| chr21_+_27303969 | 0.23 |
ENSDART00000146724
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr2_+_3125365 | 0.23 |
ENSDART00000164308
ENSDART00000167649 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr12_+_30611399 | 0.22 |
ENSDART00000153036
|
slc35f3a
|
solute carrier family 35, member F3a |
| chr25_-_18910254 | 0.22 |
ENSDART00000019765
|
efcab6
|
EF-hand calcium binding domain 6 |
| chr11_+_18710724 | 0.22 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr7_-_13661739 | 0.22 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr23_-_15090815 | 0.22 |
ENSDART00000147636
|
ndrg3b
|
ndrg family member 3b |
| chr15_-_1928150 | 0.22 |
|
|
|
| chr2_-_32404106 | 0.22 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031439 | pro-B cell differentiation(GO:0002328) regulation of mRNA cleavage(GO:0031437) positive regulation of mRNA cleavage(GO:0031439) regulation of pro-B cell differentiation(GO:2000973) positive regulation of pro-B cell differentiation(GO:2000975) |
| 0.2 | 0.6 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.2 | 0.6 | GO:0032635 | interleukin-6 production(GO:0032635) regulation of interleukin-6 production(GO:0032675) |
| 0.1 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 0.4 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 2.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.2 | GO:0030320 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.7 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.1 | 2.1 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.2 | GO:0051977 | lysophospholipid transport(GO:0051977) lipid transport across blood brain barrier(GO:1990379) |
| 0.1 | 0.8 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 1.0 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.3 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.3 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 1.4 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.4 | GO:0090316 | positive regulation of intracellular protein transport(GO:0090316) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 1.6 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 2.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.4 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.5 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.7 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 0.6 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.5 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.5 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.3 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 1.4 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.7 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 1.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 2.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 1.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 1.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 2.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.3 | GO:0016639 | glutamate dehydrogenase (NAD+) activity(GO:0004352) glutamate dehydrogenase [NAD(P)+] activity(GO:0004353) oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.9 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 1.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.2 | GO:0051978 | lysophospholipid transporter activity(GO:0051978) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.8 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.7 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |