Project
DANIO-CODE
Navigation
Downloads
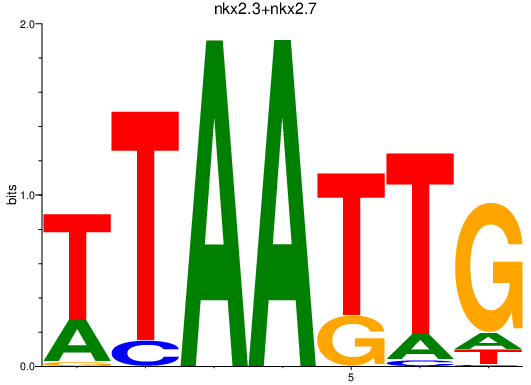
Results for nkx2.3+nkx2.7
Z-value: 1.40
Transcription factors associated with nkx2.3+nkx2.7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.7
|
ENSDARG00000021232 | NK2 transcription factor related 7 |
|
nkx2.3
|
ENSDARG00000039095 | NK2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.3 | dr10_dc_chr13_-_40286013_40286032 | 0.88 | 6.3e-06 | Click! |
| nkx2.7 | dr10_dc_chr8_-_50299273_50299328 | 0.65 | 6.0e-03 | Click! |
Activity profile of nkx2.3+nkx2.7 motif
Sorted Z-values of nkx2.3+nkx2.7 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.3+nkx2.7
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_24221087 | 3.95 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr24_+_30343717 | 3.26 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr13_-_36265504 | 3.15 |
ENSDART00000140243
|
actn1
|
actinin, alpha 1 |
| chr1_+_21037251 | 3.11 |
|
|
|
| chr23_+_19827551 | 2.96 |
ENSDART00000073442
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr17_-_29102320 | 2.87 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr12_+_14970278 | 2.87 |
ENSDART00000003847
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr17_+_23278879 | 2.75 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr1_-_58580939 | 2.73 |
ENSDART00000158011
|
col5a3b
|
collagen, type V, alpha 3b |
| chr1_+_43738350 | 2.70 |
ENSDART00000127087
ENSDART00000100309 |
crybb1l2
|
crystallin, beta B1, like 2 |
| chr21_+_28441951 | 2.65 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr5_+_31606213 | 2.64 |
ENSDART00000126873
|
myhz2
|
myosin, heavy polypeptide 2, fast muscle specific |
| chr13_+_24148599 | 2.56 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr3_-_27516974 | 2.48 |
ENSDART00000151675
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr22_-_4264838 | 2.45 |
ENSDART00000125302
|
fbn2b
|
fibrillin 2b |
| chr16_-_42040346 | 2.38 |
ENSDART00000169313
|
pycard
|
PYD and CARD domain containing |
| chr7_+_58718614 | 2.37 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr17_-_31642673 | 2.35 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr25_+_19992389 | 2.32 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr23_+_32007035 | 2.28 |
ENSDART00000132981
|
si:dkey-126g1.9
|
si:dkey-126g1.9 |
| chr13_-_13163676 | 2.28 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr10_-_17527124 | 2.28 |
ENSDART00000137905
|
nt5c2l1
|
5'-nucleotidase, cytosolic II, like 1 |
| chr24_+_2523708 | 2.27 |
ENSDART00000106619
|
nrn1a
|
neuritin 1a |
| chr8_-_23172938 | 2.13 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr4_+_12932838 | 2.12 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr15_-_35563367 | 2.11 |
|
|
|
| chr2_-_20941256 | 2.07 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr1_-_35419335 | 2.05 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr24_+_30343687 | 1.97 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr14_+_20045365 | 1.97 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr17_-_52735250 | 1.96 |
|
|
|
| chr20_+_2443682 | 1.91 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr6_-_23455346 | 1.90 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr11_+_41401893 | 1.89 |
ENSDART00000171655
|
AL929185.1
|
ENSDARG00000103982 |
| chr21_+_7844259 | 1.89 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr13_+_22350043 | 1.86 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr1_-_37475807 | 1.86 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr2_-_30675594 | 1.85 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr5_-_41672394 | 1.84 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr15_-_15421065 | 1.81 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr5_-_15774816 | 1.78 |
ENSDART00000090684
|
kremen1
|
kringle containing transmembrane protein 1 |
| chr22_-_37417903 | 1.77 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr18_-_3056732 | 1.76 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr1_-_37990863 | 1.75 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr19_-_29205158 | 1.75 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr24_+_9605832 | 1.74 |
ENSDART00000131891
|
tmem108
|
transmembrane protein 108 |
| chr13_-_36265476 | 1.73 |
ENSDART00000133740
ENSDART00000100217 |
actn1
|
actinin, alpha 1 |
| chr6_+_23787804 | 1.69 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr6_-_8075384 | 1.68 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr22_-_22846670 | 1.68 |
ENSDART00000176355
|
CABZ01039424.2
|
ENSDARG00000108442 |
| chr5_-_58269742 | 1.67 |
ENSDART00000122413
|
mcama
|
melanoma cell adhesion molecule a |
| chr21_-_43020159 | 1.65 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr20_+_28958586 | 1.63 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr16_-_17439735 | 1.63 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr4_+_8531280 | 1.61 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr2_+_25363717 | 1.61 |
ENSDART00000142601
|
stag1a
|
stromal antigen 1a |
| chr18_+_48426375 | 1.57 |
|
|
|
| chr1_-_4757890 | 1.56 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr6_+_28886741 | 1.55 |
ENSDART00000065137
|
tp63
|
tumor protein p63 |
| chr5_+_37254520 | 1.53 |
ENSDART00000051222
|
ins
|
preproinsulin |
| chr16_+_14817718 | 1.53 |
ENSDART00000027982
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr2_-_36058327 | 1.51 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr14_-_5371581 | 1.51 |
ENSDART00000012116
|
tlx2
|
T-cell leukemia, homeobox 2 |
| chr12_+_34852049 | 1.50 |
ENSDART00000027034
|
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr13_-_13163801 | 1.49 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr5_-_47523737 | 1.48 |
ENSDART00000153239
|
BX465834.1
|
ENSDARG00000095715 |
| chr13_-_44492897 | 1.48 |
|
|
|
| chr3_+_23557320 | 1.48 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr9_-_33584190 | 1.47 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr7_+_34018862 | 1.46 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr25_-_18855316 | 1.46 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr12_-_6144233 | 1.45 |
ENSDART00000152292
|
a1cf
|
apobec1 complementation factor |
| chr19_-_8829565 | 1.44 |
ENSDART00000031173
|
rps27.1
|
ribosomal protein S27, isoform 1 |
| chr17_+_52736192 | 1.43 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr1_+_10036098 | 1.42 |
ENSDART00000139387
|
atp1b1b
|
ATPase, Na+/K+ transporting, beta 1b polypeptide |
| chr2_+_21067708 | 1.42 |
ENSDART00000148400
ENSDART00000021168 |
rxrga
|
retinoid x receptor, gamma a |
| chr2_-_32370176 | 1.41 |
ENSDART00000077151
|
asap1a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1a |
| chr2_-_59019569 | 1.40 |
|
|
|
| chr3_-_13942505 | 1.40 |
|
|
|
| chr10_-_8074713 | 1.39 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase, H+ transporting, lysosomal V0 subunit a2a |
| chr13_+_30781652 | 1.39 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr8_-_31065717 | 1.38 |
ENSDART00000163407
|
slc20a1a
|
solute carrier family 20, member 1a |
| chr14_+_14262816 | 1.38 |
ENSDART00000164749
|
pcdh20
|
protocadherin 20 |
| chr3_+_23638277 | 1.38 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr9_+_21911860 | 1.38 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr1_-_10391257 | 1.37 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr9_-_14533551 | 1.34 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr13_+_24531753 | 1.34 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr5_+_44407763 | 1.34 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr16_-_36022327 | 1.31 |
ENSDART00000172252
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr9_+_48717490 | 1.30 |
ENSDART00000163353
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr3_-_32464649 | 1.30 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr2_-_10555152 | 1.30 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr2_+_32036450 | 1.29 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr18_+_48959401 | 1.27 |
ENSDART00000158104
|
SHKBP1
|
SH3KBP1 binding protein 1 |
| chr12_+_25684420 | 1.27 |
ENSDART00000024415
|
epas1a
|
endothelial PAS domain protein 1a |
| chr12_+_32936230 | 1.27 |
ENSDART00000153146
|
rbfox3a
|
RNA binding protein, fox-1 homolog (C. elegans) 3a |
| chr9_+_30767182 | 1.27 |
ENSDART00000139811
|
commd6
|
COMM domain containing 6 |
| chr13_+_23065500 | 1.25 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr19_-_2486568 | 1.25 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| KN149858v1_-_12281 | 1.25 |
ENSDART00000157456
|
CABZ01056637.1
|
ENSDARG00000100203 |
| chr12_-_30897542 | 1.24 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr2_+_25659945 | 1.24 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr21_-_9863186 | 1.24 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr12_-_37921731 | 1.23 |
|
|
|
| chr17_-_52735615 | 1.22 |
|
|
|
| chr7_+_32451041 | 1.22 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr13_+_28689749 | 1.22 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr8_-_22945616 | 1.22 |
|
|
|
| chr2_-_20490037 | 1.22 |
ENSDART00000160388
|
FQ377605.1
|
ENSDARG00000101927 |
| chr8_-_50159285 | 1.21 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr7_+_47014195 | 1.21 |
ENSDART00000114669
|
dpy19l3
|
dpy-19-like 3 (C. elegans) |
| chr12_-_25825072 | 1.21 |
|
|
|
| chr14_-_24113324 | 1.20 |
ENSDART00000125923
|
cxcl14
|
chemokine (C-X-C motif) ligand 14 |
| chr5_+_36431824 | 1.18 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr24_-_16994956 | 1.17 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr1_-_11286438 | 1.16 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr3_-_61104221 | 1.16 |
ENSDART00000155414
|
tecpr1b
|
tectonin beta-propeller repeat containing 1b |
| chr1_-_12584282 | 1.16 |
ENSDART00000127838
|
pcdh18a
|
protocadherin 18a |
| chr18_+_23891068 | 1.16 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr6_+_3771586 | 1.15 |
|
|
|
| chr21_-_20305406 | 1.15 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr25_+_30701751 | 1.15 |
|
|
|
| chr5_+_62987426 | 1.14 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr8_-_15091823 | 1.14 |
ENSDART00000142358
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr20_-_23272102 | 1.13 |
ENSDART00000167197
|
spata18
|
spermatogenesis associated 18 |
| chr14_-_9216303 | 1.12 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr8_+_23716843 | 1.12 |
ENSDART00000136547
|
rpl10a
|
ribosomal protein L10a |
| chr3_+_3955069 | 1.12 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr21_+_28921734 | 1.11 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_32942783 | 1.11 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr8_+_14120313 | 1.11 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr22_+_16279764 | 1.11 |
|
|
|
| chr13_+_19191645 | 1.10 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr15_+_43714699 | 1.10 |
ENSDART00000168191
|
ENSDARG00000102235
|
ENSDARG00000102235 |
| chr16_+_21104310 | 1.10 |
ENSDART00000046766
|
hoxa10b
|
homeobox A10b |
| chr3_-_39554155 | 1.09 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr14_-_40918015 | 1.08 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr9_+_51733862 | 1.08 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr6_+_8895437 | 1.08 |
ENSDART00000162588
|
rgn
|
regucalcin |
| chr5_-_67233396 | 1.07 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr11_-_1409262 | 1.07 |
ENSDART00000155269
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr1_+_529491 | 1.07 |
ENSDART00000102421
|
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr17_+_24613874 | 1.06 |
ENSDART00000093004
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr19_+_41488385 | 1.05 |
ENSDART00000138687
|
ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr19_-_6856033 | 1.05 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr16_-_6450531 | 1.05 |
ENSDART00000060550
ENSDART00000145663 |
ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr24_-_8589270 | 1.04 |
ENSDART00000082346
|
tfap2a
|
transcription factor AP-2 alpha |
| chr10_-_43466392 | 1.04 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr18_-_41384938 | 1.04 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr7_-_25816549 | 1.03 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr13_+_30781718 | 1.03 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr3_-_27517052 | 1.03 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr17_-_44133941 | 1.02 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr16_+_26576242 | 1.02 |
ENSDART00000039746
|
epb41b
|
erythrocyte membrane protein band 4.1b |
| chr11_-_27253835 | 1.02 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr13_+_28286826 | 0.99 |
ENSDART00000043658
|
slc2a15a
|
solute carrier family 2 (facilitated glucose transporter), member 15a |
| chr10_-_43466478 | 0.98 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr19_-_42821964 | 0.98 |
ENSDART00000087002
|
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr7_-_71638662 | 0.98 |
ENSDART00000162984
|
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr5_-_31301280 | 0.98 |
ENSDART00000141446
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr5_-_10445847 | 0.97 |
ENSDART00000163139
|
rtn4r
|
reticulon 4 receptor |
| chr20_+_33972327 | 0.97 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr9_+_25964868 | 0.97 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr13_-_44492701 | 0.97 |
|
|
|
| chr10_+_17494274 | 0.96 |
|
|
|
| chr21_+_35181329 | 0.96 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr7_-_39170016 | 0.95 |
ENSDART00000161191
|
BX842701.1
|
ENSDARG00000101872 |
| chr17_+_29259577 | 0.95 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr4_+_12932950 | 0.95 |
ENSDART00000016382
|
wif1
|
wnt inhibitory factor 1 |
| chr15_-_34354293 | 0.95 |
|
|
|
| chr2_+_3370130 | 0.95 |
ENSDART00000098391
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr24_+_24924379 | 0.93 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr19_+_2754903 | 0.93 |
ENSDART00000160533
|
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr17_-_21046246 | 0.92 |
ENSDART00000078763
|
vsx1
|
visual system homeobox 1 homolog, chx10-like |
| chr15_-_20088439 | 0.92 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr1_+_30992553 | 0.91 |
ENSDART00000112333
|
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr9_-_37940101 | 0.90 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr4_-_2549493 | 0.90 |
|
|
|
| chr24_-_6048914 | 0.90 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr6_-_7563110 | 0.90 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr13_+_24703802 | 0.90 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr16_-_37461878 | 0.89 |
ENSDART00000142916
|
si:ch211-208k15.1
|
si:ch211-208k15.1 |
| chr1_-_50215233 | 0.88 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr16_+_10531926 | 0.88 |
ENSDART00000161568
|
ino80e
|
INO80 complex subunit E |
| chr12_+_5046825 | 0.88 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr6_-_23455458 | 0.88 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr9_-_23307419 | 0.88 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr24_-_6129575 | 0.87 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr18_+_23891152 | 0.87 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr6_+_45493396 | 0.87 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| chr19_-_4079427 | 0.87 |
ENSDART00000170325
|
map7d1b
|
MAP7 domain containing 1b |
| chr5_-_13826859 | 0.86 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr20_-_37587804 | 0.86 |
ENSDART00000166106
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr23_+_2193582 | 0.85 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr24_-_16995194 | 0.85 |
ENSDART00000111079
|
CR376737.1
|
ENSDARG00000077318 |
| chr8_+_20456215 | 0.85 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr21_-_7617151 | 0.85 |
ENSDART00000170166
|
fam69b
|
family with sequence similarity 69, member B |
| chr21_-_19293600 | 0.84 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr15_+_4079643 | 0.84 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:2000136 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.8 | 3.4 | GO:0035908 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.8 | 2.3 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.6 | 1.9 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.6 | 1.7 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.5 | 1.5 | GO:0006581 | response to amphetamine(GO:0001975) acetylcholine catabolic process(GO:0006581) response to amine(GO:0014075) |
| 0.5 | 1.4 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.4 | 0.9 | GO:0050886 | endocrine process(GO:0050886) |
| 0.4 | 1.2 | GO:0051701 | interaction with host(GO:0051701) |
| 0.4 | 1.5 | GO:0060259 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of feeding behavior(GO:0060259) |
| 0.4 | 7.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 1.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.4 | 1.8 | GO:0048903 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.3 | 1.7 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.3 | 1.4 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.3 | 1.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 0.9 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.3 | 1.2 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.3 | 1.2 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 0.6 | GO:0097378 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.3 | 3.6 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 4.1 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.3 | 1.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.3 | 1.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.3 | 1.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.3 | 1.0 | GO:0019343 | homoserine metabolic process(GO:0009092) cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) transsulfuration(GO:0019346) |
| 0.2 | 1.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 0.7 | GO:0030818 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.2 | 0.7 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.1 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 1.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.2 | 0.8 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.2 | 0.6 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 0.8 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 2.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 0.6 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.2 | 2.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.2 | 0.6 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.2 | 1.8 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.2 | 0.5 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.2 | 1.0 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.2 | 0.6 | GO:1901389 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.2 | 1.9 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.2 | 3.0 | GO:0014823 | response to activity(GO:0014823) |
| 0.2 | 1.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 1.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.9 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 1.8 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 1.5 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.8 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.1 | 0.9 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 2.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.4 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 1.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.5 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 1.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.1 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 0.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.1 | 1.5 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.1 | 4.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 2.1 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 2.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.9 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 2.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.9 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 2.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 2.7 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.1 | 1.0 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 2.9 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 1.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 1.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 1.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0032096 | regulation of response to food(GO:0032095) negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 2.4 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.2 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.1 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.5 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.1 | 0.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.2 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.7 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.1 | 0.3 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.2 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 1.7 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.5 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 2.6 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 5.8 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 4.4 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.3 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 1.2 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.0 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.1 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.4 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 0.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 1.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 1.0 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 1.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 1.8 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.8 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 1.6 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 1.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.1 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.0 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.8 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 4.1 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.4 | 2.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.1 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 0.7 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 2.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.2 | 1.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.4 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 1.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 6.7 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 2.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 5.1 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 5.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 1.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.6 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.6 | GO:0042645 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.0 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 4.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 4.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.7 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 8.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 0.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 10.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.0 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.8 | 2.4 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 1.6 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 1.5 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.5 | 2.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.0 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 1.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 0.9 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 1.4 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.3 | 2.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 1.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.3 | 4.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.7 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 1.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.8 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.2 | 1.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.2 | 0.7 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.2 | 1.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 0.6 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 1.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.6 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.4 | GO:0016175 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 1.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.5 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.0 | GO:0042379 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.1 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 5.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 2.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 3.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.5 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.1 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 2.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 0.8 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.2 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.3 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 7.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 1.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 1.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 6.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 4.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 2.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 22.2 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 3.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 5.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.2 | 4.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.2 | 3.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 4.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 3.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 3.0 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.3 | 4.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.3 | 3.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 1.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 1.4 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 2.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 1.6 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 0.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |