Project
DANIO-CODE
Navigation
Downloads
Results for nkx2.4a+nkx2.4b
Z-value: 0.86
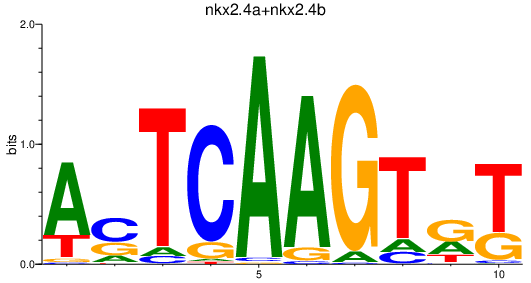
Transcription factors associated with nkx2.4a+nkx2.4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.4a
|
ENSDARG00000075107 | NK2 homeobox 4a |
|
nkx2.4b
|
ENSDARG00000104107 | NK2 homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx2.4a | dr10_dc_chr17_-_42108276_42108374 | 0.82 | 1.1e-04 | Click! |
| nkx2.4b | dr10_dc_chr20_+_48971948_48971982 | 0.81 | 1.6e-04 | Click! |
Activity profile of nkx2.4a+nkx2.4b motif
Sorted Z-values of nkx2.4a+nkx2.4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.4a+nkx2.4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_13163676 | 2.12 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr2_+_42874975 | 2.09 |
ENSDART00000075392
|
basp1
|
brain abundant, membrane attached signal protein 1 |
| chr14_-_26238386 | 1.91 |
ENSDART00000105933
|
tgfbi
|
transforming growth factor, beta-induced |
| chr7_+_25052543 | 1.89 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr1_+_25005968 | 1.84 |
ENSDART00000054228
|
lrata
|
lecithin retinol acyltransferase a (phosphatidylcholine-retinol O-acyltransferase) |
| chr8_+_23464087 | 1.76 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr10_-_10059864 | 1.63 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr10_+_44364117 | 1.63 |
|
|
|
| chr19_-_23037220 | 1.59 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr25_-_27399421 | 1.52 |
ENSDART00000156906
|
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr17_+_27417635 | 1.49 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr10_+_21850059 | 1.46 |
ENSDART00000164634
ENSDART00000172513 |
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr16_+_22950567 | 1.42 |
ENSDART00000143957
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr5_+_36010448 | 1.38 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr8_-_49356479 | 1.38 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr13_-_13163801 | 1.36 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr14_+_13919450 | 1.26 |
ENSDART00000174760
|
nlgn3a
|
neuroligin 3a |
| chr23_+_14058972 | 1.23 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr5_-_33225263 | 1.16 |
ENSDART00000139371
|
BX005445.1
|
ENSDARG00000095064 |
| chr24_+_30343717 | 1.15 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr14_-_32877752 | 1.14 |
ENSDART00000163046
|
si:dkey-31j3.11
|
si:dkey-31j3.11 |
| chr22_+_12746080 | 1.13 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr21_-_41286846 | 1.13 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr21_-_39132283 | 1.12 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr15_+_9096765 | 1.11 |
ENSDART00000080196
|
si:dkey-202g17.3
|
si:dkey-202g17.3 |
| chr23_+_36002332 | 1.07 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr22_-_36915567 | 1.03 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr8_-_17480730 | 0.99 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr23_-_7892862 | 0.99 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr10_-_32580145 | 0.98 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr7_+_25052687 | 0.97 |
ENSDART00000110347
|
cyp26b1
|
cytochrome P450, family 26, subfamily b, polypeptide 1 |
| chr13_+_30546398 | 0.94 |
|
|
|
| chr10_+_31358236 | 0.94 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr9_-_12840621 | 0.94 |
ENSDART00000088042
|
myo10l3
|
myosin X-like 3 |
| chr5_+_53178362 | 0.93 |
ENSDART00000169565
|
sat2b
|
spermidine/spermine N1-acetyltransferase family member 2b |
| chr11_-_30105047 | 0.92 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr5_+_32282547 | 0.88 |
ENSDART00000135459
|
rpl12
|
ribosomal protein L12 |
| chr20_-_38546922 | 0.88 |
ENSDART00000136303
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr20_+_27565701 | 0.86 |
|
|
|
| chr24_+_35899507 | 0.84 |
ENSDART00000122408
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr19_+_24398194 | 0.84 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr17_+_43041951 | 0.84 |
ENSDART00000154863
|
isca2
|
iron-sulfur cluster assembly 2 |
| chr2_+_393439 | 0.83 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr6_+_37881015 | 0.81 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr14_+_23412305 | 0.80 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr8_-_24276303 | 0.80 |
|
|
|
| chr3_-_29992106 | 0.80 |
ENSDART00000153562
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr7_-_34854886 | 0.78 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr19_-_28192439 | 0.76 |
ENSDART00000135348
|
adcy2b
|
adenylate cyclase 2b (brain) |
| chr18_-_47186938 | 0.73 |
|
|
|
| chr4_+_18974767 | 0.69 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr9_+_7745593 | 0.67 |
ENSDART00000061716
|
mnx2a
|
motor neuron and pancreas homeobox 2a |
| chr14_+_20809272 | 0.67 |
ENSDART00000139865
|
aldob
|
aldolase b, fructose-bisphosphate |
| chr10_-_32580373 | 0.65 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr13_-_1980818 | 0.65 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr2_-_49248076 | 0.64 |
ENSDART00000161156
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr16_-_34304851 | 0.63 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr12_-_31342432 | 0.61 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr13_+_26943081 | 0.60 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr15_-_12079779 | 0.60 |
ENSDART00000160427
|
si:dkey-202l22.6
|
si:dkey-202l22.6 |
| chr11_-_29908609 | 0.59 |
ENSDART00000156196
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr10_-_15090647 | 0.59 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr24_+_30343687 | 0.58 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr2_+_16983582 | 0.57 |
ENSDART00000091351
|
gk5
|
glycerol kinase 5 (putative) |
| chr24_+_35173816 | 0.54 |
|
|
|
| chr14_-_51867502 | 0.52 |
ENSDART00000166924
|
ENSDARG00000098660
|
ENSDARG00000098660 |
| chr6_+_3703684 | 0.51 |
ENSDART00000013743
|
gorasp2
|
golgi reassembly stacking protein 2 |
| chr5_+_34381412 | 0.51 |
ENSDART00000134795
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr9_-_23222916 | 0.49 |
ENSDART00000022392
ENSDART00000122036 |
rnd3b
|
Rho family GTPase 3b |
| chr6_+_46968199 | 0.48 |
|
|
|
| chr6_+_37880936 | 0.48 |
ENSDART00000087311
|
oca2
|
oculocutaneous albinism II |
| chr25_+_1773157 | 0.47 |
|
|
|
| chr3_+_33209227 | 0.47 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr24_-_16993914 | 0.45 |
|
|
|
| chr13_-_13163647 | 0.45 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr17_-_42778156 | 0.41 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr11_+_35896325 | 0.40 |
ENSDART00000163330
|
grm2b
|
glutamate receptor, metabotropic 2b |
| chr18_-_47186896 | 0.38 |
|
|
|
| chr11_+_38858351 | 0.37 |
ENSDART00000155746
|
cdc42
|
cell division cycle 42 |
| chr20_+_48950833 | 0.37 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr18_+_29166912 | 0.37 |
ENSDART00000089031
|
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr6_+_58837900 | 0.36 |
ENSDART00000157581
|
dctn2
|
dynactin 2 (p50) |
| chr8_+_21556157 | 0.36 |
ENSDART00000172190
|
ajap1
|
adherens junctions associated protein 1 |
| chr1_-_33880029 | 0.36 |
ENSDART00000004361
|
klf5a
|
Kruppel-like factor 5a |
| chr25_-_12633686 | 0.35 |
ENSDART00000171144
ENSDART00000171834 |
klhdc4
|
kelch domain containing 4 |
| chr6_-_26897413 | 0.35 |
|
|
|
| chr13_-_30807799 | 0.34 |
|
|
|
| chr3_+_24003840 | 0.34 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr10_+_44364064 | 0.33 |
|
|
|
| chr7_-_33080261 | 0.33 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr9_+_2756231 | 0.33 |
ENSDART00000123342
|
sp3a
|
sp3a transcription factor |
| chr20_-_26973962 | 0.32 |
|
|
|
| chr10_+_31358307 | 0.31 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr20_+_27121126 | 0.30 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr18_+_20058028 | 0.30 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr22_+_39110524 | 0.29 |
ENSDART00000153841
|
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr23_-_18972097 | 0.28 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr2_-_11864995 | 0.28 |
ENSDART00000080925
|
impad1
|
inositol monophosphatase domain containing 1 |
| chr5_-_32282404 | 0.28 |
ENSDART00000166698
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr2_-_6380457 | 0.27 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr20_+_23843436 | 0.27 |
ENSDART00000050559
|
sh3rf1
|
SH3 domain containing ring finger 1 |
| chr13_+_2213363 | 0.27 |
ENSDART00000019989
ENSDART00000165305 |
fbxo9
|
F-box protein 9 |
| chr19_-_6215704 | 0.26 |
ENSDART00000104978
|
cica
|
capicua transcriptional repressor a |
| chr2_-_6380589 | 0.26 |
ENSDART00000092182
|
ppm1la
|
protein phosphatase, Mg2+/Mn2+ dependent, 1La |
| chr17_+_33388313 | 0.25 |
ENSDART00000077553
|
xdh
|
xanthine dehydrogenase |
| chr3_+_60367511 | 0.24 |
ENSDART00000128260
|
CABZ01087514.1
|
ENSDARG00000090861 |
| chr5_-_4082749 | 0.23 |
|
|
|
| chr6_+_12999420 | 0.23 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr12_-_48278273 | 0.22 |
|
|
|
| chr18_+_92690 | 0.21 |
|
|
|
| chr5_-_32860888 | 0.20 |
ENSDART00000085636
|
ENSDARG00000060631
|
ENSDARG00000060631 |
| chr24_-_10910835 | 0.20 |
ENSDART00000133530
|
asap1b
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1b |
| chr7_-_18348841 | 0.19 |
ENSDART00000111636
|
DTX4
|
deltex E3 ubiquitin ligase 4 |
| chr15_-_34700453 | 0.14 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr19_-_38832599 | 0.14 |
ENSDART00000087639
|
smap2
|
small ArfGAP2 |
| chr23_+_34392753 | 0.14 |
|
|
|
| chr22_-_36915689 | 0.13 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr7_-_6222134 | 0.13 |
ENSDART00000173197
|
hist1h4l
|
histone 1, H4, like |
| chr10_-_28638101 | 0.12 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr13_-_30807601 | 0.11 |
|
|
|
| chr6_-_59412311 | 0.11 |
|
|
|
| chr21_-_2322963 | 0.10 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr24_+_33478309 | 0.10 |
ENSDART00000122579
|
si:ch73-173p19.1
|
si:ch73-173p19.1 |
| chr20_-_51585893 | 0.09 |
ENSDART00000151426
|
si:ch73-91k6.2
|
si:ch73-91k6.2 |
| chr24_+_3177066 | 0.09 |
|
|
|
| chr23_-_33783396 | 0.08 |
ENSDART00000130338
|
pou6f1
|
POU class 6 homeobox 1 |
| chr17_-_31027949 | 0.08 |
ENSDART00000152016
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr11_+_21949272 | 0.08 |
ENSDART00000122136
|
MDFI
|
MyoD family inhibitor |
| chr20_-_26973934 | 0.07 |
|
|
|
| chr7_-_41871376 | 0.04 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr2_-_6380096 | 0.04 |
|
|
|
| chr2_-_38330046 | 0.04 |
ENSDART00000076503
|
ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr19_-_38832891 | 0.03 |
ENSDART00000087639
|
smap2
|
small ArfGAP2 |
| chr9_+_41776562 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.5 | 1.6 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.4 | 1.3 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.4 | 1.1 | GO:0052803 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.4 | 1.4 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 1.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 0.9 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.3 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.2 | 0.9 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 1.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.6 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 1.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 1.2 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.2 | 0.8 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 3.9 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.8 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 0.3 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.1 | 0.5 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.1 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.9 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.7 | GO:0006183 | GMP biosynthetic process(GO:0006177) GTP biosynthetic process(GO:0006183) GMP metabolic process(GO:0046037) |
| 0.1 | 1.1 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.4 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.3 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.5 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 1.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.1 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 1.9 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.3 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 1.6 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.6 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 1.1 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 1.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 1.6 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 10.0 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.0 | 1.9 | GO:0031012 | extracellular matrix(GO:0031012) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 1.1 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.4 | 1.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.3 | 0.8 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 3.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.9 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 0.7 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.2 | 1.6 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.2 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.2 | 0.9 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.2 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.1 | 0.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.9 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.3 | GO:0030151 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 1.8 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.8 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.3 | GO:0003774 | motor activity(GO:0003774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.9 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 2.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.9 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.1 | 2.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.2 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 2.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |