Project
DANIO-CODE
Navigation
Downloads
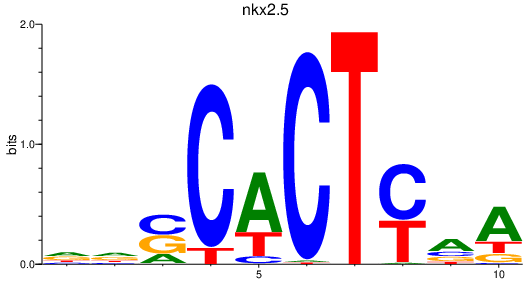
Results for nkx2.5
Z-value: 0.96
Transcription factors associated with nkx2.5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx2.5
|
ENSDARG00000018004 | NK2 homeobox 5 |
Activity profile of nkx2.5 motif
Sorted Z-values of nkx2.5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx2.5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_4567653 | 3.48 |
|
|
|
| chr15_+_29153215 | 2.61 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr25_-_14472107 | 2.05 |
|
|
|
| chr1_+_49894978 | 1.96 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr20_+_4567603 | 1.79 |
|
|
|
| chr15_-_18425091 | 1.73 |
ENSDART00000155866
|
zbtb16b
|
zinc finger and BTB domain containing 16b |
| chr3_+_32394653 | 1.69 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr8_+_48106836 | 1.67 |
ENSDART00000138717
|
si:ch211-263k4.2
|
si:ch211-263k4.2 |
| chr5_-_42272455 | 1.65 |
ENSDART00000161586
|
flot2a
|
flotillin 2a |
| chr23_+_19827551 | 1.65 |
ENSDART00000073442
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr18_-_46243753 | 1.64 |
ENSDART00000145999
|
prx
|
periaxin |
| chr1_+_26330186 | 1.43 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr10_-_39210658 | 1.43 |
ENSDART00000150193
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr4_-_24310846 | 1.42 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr16_-_46653204 | 1.26 |
ENSDART00000158341
|
tmem176l.3a
|
transmembrane protein 176l.3a |
| chr20_+_2443682 | 1.26 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr16_-_9785057 | 1.25 |
ENSDART00000113724
|
mal2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr7_-_47900199 | 1.25 |
ENSDART00000174262
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr14_+_21531709 | 1.24 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr15_-_47277391 | 1.23 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr5_-_248845 | 1.20 |
ENSDART00000127504
|
trabd2a
|
TraB domain containing 2A |
| chr23_-_35657313 | 1.18 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| KN150131v1_+_59098 | 1.17 |
ENSDART00000175590
|
CABZ01064941.1
|
ENSDARG00000106550 |
| chr14_-_48951428 | 1.16 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr9_+_30483319 | 1.16 |
ENSDART00000148148
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr12_+_6065116 | 1.16 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr14_-_7774262 | 1.13 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr12_-_17357133 | 1.13 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr6_-_39346614 | 1.13 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr24_-_2918801 | 1.12 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr12_+_2835894 | 1.12 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr21_-_13571753 | 1.10 |
ENSDART00000065819
|
clic3
|
chloride intracellular channel 3 |
| chr13_-_49826330 | 1.08 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr17_+_45824480 | 1.07 |
|
|
|
| chr9_-_12726136 | 1.06 |
|
|
|
| chr22_+_5657904 | 1.05 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr4_+_21996784 | 1.05 |
ENSDART00000040266
|
myf6
|
myogenic factor 6 |
| chr21_-_28486800 | 1.05 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr11_+_37639045 | 1.02 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr11_-_33697442 | 1.01 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr3_+_22246697 | 1.01 |
ENSDART00000155597
|
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr10_+_5693933 | 0.97 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_25272318 | 0.97 |
ENSDART00000134192
|
synpo2b
|
synaptopodin 2b |
| chr5_-_70939686 | 0.96 |
|
|
|
| chr18_+_18598885 | 0.95 |
|
|
|
| chr20_-_28739099 | 0.94 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr9_-_22347397 | 0.92 |
ENSDART00000146188
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr22_+_15946773 | 0.89 |
ENSDART00000144545
|
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr9_-_35135414 | 0.89 |
ENSDART00000140563
|
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr3_-_7220340 | 0.88 |
|
|
|
| chr16_-_24696036 | 0.87 |
|
|
|
| chr20_+_23599157 | 0.87 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr24_-_2278409 | 0.86 |
|
|
|
| chr15_-_41288480 | 0.85 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr25_-_31881585 | 0.84 |
ENSDART00000123999
ENSDART00000158091 |
ENSDARG00000100731
|
ENSDARG00000100731 |
| chr1_-_44942144 | 0.84 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr17_-_44135924 | 0.83 |
ENSDART00000156648
|
otx2
|
orthodenticle homeobox 2 |
| chr20_+_44413431 | 0.82 |
ENSDART00000114660
|
opn8b
|
opsin 8, group member b |
| chr6_+_24717985 | 0.81 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr18_+_46384462 | 0.80 |
ENSDART00000024202
|
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr10_-_24401876 | 0.79 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr13_+_22134507 | 0.78 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr1_-_44942047 | 0.76 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr7_-_32604392 | 0.76 |
ENSDART00000132504
|
ano5b
|
anoctamin 5b |
| chr22_+_18364282 | 0.76 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr6_-_22869821 | 0.74 |
ENSDART00000167674
|
mif4gda
|
MIF4G domain containing a |
| chr1_+_49894685 | 0.73 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr21_+_45693978 | 0.71 |
ENSDART00000160324
|
CABZ01102047.1
|
ENSDARG00000099648 |
| chr3_-_33295814 | 0.71 |
ENSDART00000146294
|
rpl23
|
ribosomal protein L23 |
| chr22_+_1320235 | 0.70 |
|
|
|
| chr22_+_1404309 | 0.70 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr17_-_610651 | 0.70 |
|
|
|
| chr14_-_205130 | 0.69 |
ENSDART00000035581
ENSDART00000158725 ENSDART00000163549 |
OTOP1
|
otopetrin 1 |
| chr2_-_43997672 | 0.69 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| KN149830v1_-_23361 | 0.68 |
|
|
|
| chr6_-_40660116 | 0.68 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr12_+_22455344 | 0.67 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr19_+_46647893 | 0.65 |
ENSDART00000162785
ENSDART00000171251 ENSDART00000164938 |
mapk15
|
mitogen-activated protein kinase 15 |
| chr13_-_7897936 | 0.63 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr18_+_41243277 | 0.63 |
|
|
|
| chr25_-_10919875 | 0.63 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
| chr25_+_35602451 | 0.61 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr16_+_9718837 | 0.60 |
ENSDART00000047920
|
enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr10_+_2952793 | 0.58 |
|
|
|
| chr19_+_26838851 | 0.57 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr16_-_2570425 | 0.57 |
|
|
|
| chr19_-_31785558 | 0.57 |
ENSDART00000046609
|
scin
|
scinderin |
| chr22_-_337198 | 0.57 |
|
|
|
| chr4_-_13903253 | 0.55 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr7_-_47900141 | 0.55 |
ENSDART00000174262
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr11_-_3609813 | 0.55 |
ENSDART00000136577
|
itih3a
|
inter-alpha-trypsin inhibitor heavy chain 3a |
| chr7_-_17129072 | 0.55 |
|
|
|
| chr7_+_40367221 | 0.54 |
ENSDART00000052236
|
mnx1
|
motor neuron and pancreas homeobox 1 |
| chr15_-_41777233 | 0.53 |
ENSDART00000154230
|
ftr90
|
finTRIM family, member 90 |
| chr8_-_38316609 | 0.53 |
|
|
|
| chr1_+_26330335 | 0.51 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr16_+_55008251 | 0.51 |
ENSDART00000176995
|
CABZ01087388.1
|
ENSDARG00000106538 |
| chr1_-_30772653 | 0.51 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr23_+_4385722 | 0.50 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr22_+_38206685 | 0.47 |
ENSDART00000104527
|
tm4sf18
|
transmembrane 4 L six family member 18 |
| chr21_-_39900518 | 0.47 |
ENSDART00000175055
|
CABZ01065291.1
|
ENSDARG00000107374 |
| chr16_+_24046789 | 0.46 |
|
|
|
| chr1_+_50348973 | 0.46 |
|
|
|
| chr9_-_2416300 | 0.46 |
|
|
|
| chr11_+_38013564 | 0.45 |
ENSDART00000171496
|
CDK18
|
cyclin dependent kinase 18 |
| chr1_-_31117154 | 0.44 |
|
|
|
| chr13_-_50299821 | 0.44 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr11_-_16067646 | 0.43 |
|
|
|
| chr8_+_17803229 | 0.42 |
ENSDART00000175436
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr23_-_28367816 | 0.41 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr4_-_4110484 | 0.41 |
ENSDART00000138221
|
lmod2b
|
leiomodin 2 (cardiac) b |
| chr6_+_58837900 | 0.40 |
ENSDART00000157581
|
dctn2
|
dynactin 2 (p50) |
| chr4_-_74789596 | 0.39 |
ENSDART00000174356
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr5_+_68352513 | 0.39 |
ENSDART00000160775
|
ENSDARG00000102513
|
ENSDARG00000102513 |
| chr18_-_43890514 | 0.39 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr7_-_68033217 | 0.38 |
|
|
|
| chr16_-_31868810 | 0.36 |
ENSDART00000058737
|
cdc42l
|
cell division cycle 42, like |
| chr14_-_48951396 | 0.36 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr12_-_7790309 | 0.36 |
ENSDART00000148673
|
ank3b
|
ankyrin 3b |
| chr18_-_3030681 | 0.35 |
ENSDART00000161848
|
ints4
|
integrator complex subunit 4 |
| chr3_+_52746124 | 0.35 |
|
|
|
| chr3_+_32394550 | 0.34 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr24_-_33361897 | 0.34 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr11_-_25222937 | 0.34 |
|
|
|
| chr3_+_19582280 | 0.34 |
ENSDART00000109409
|
mrc2
|
mannose receptor, C type 2 |
| chr1_+_46487300 | 0.34 |
ENSDART00000158432
ENSDART00000074450 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr1_+_33510660 | 0.34 |
ENSDART00000147201
|
slc5a7a
|
solute carrier family 5 (sodium/choline cotransporter), member 7a |
| chr5_+_31562242 | 0.33 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr20_+_27068108 | 0.33 |
ENSDART00000132434
ENSDART00000029781 |
ahsa1a
|
AHA1, activator of heat shock protein ATPase homolog 1a |
| chr13_-_49920975 | 0.33 |
ENSDART00000165066
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr11_+_25266623 | 0.32 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr13_-_50299643 | 0.31 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr15_+_24277664 | 0.31 |
ENSDART00000155502
|
sez6b
|
seizure related 6 homolog b |
| chr10_+_22043094 | 0.30 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr1_-_54593214 | 0.29 |
ENSDART00000152807
|
si:ch211-286b5.5
|
si:ch211-286b5.5 |
| chr2_-_24735186 | 0.29 |
ENSDART00000141365
|
zgc:154006
|
zgc:154006 |
| chr15_-_1519571 | 0.28 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr13_-_6089739 | 0.28 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr6_-_45867149 | 0.27 |
ENSDART00000062459
|
rbm19
|
RNA binding motif protein 19 |
| chr6_-_25962268 | 0.27 |
ENSDART00000076997
|
lmo4b
|
LIM domain only 4b |
| chr22_-_12130441 | 0.25 |
ENSDART00000146785
|
tmem163b
|
transmembrane protein 163b |
| chr14_-_32956886 | 0.25 |
ENSDART00000075056
|
rab41
|
RAB41, member RAS oncogene family |
| chr8_+_25594106 | 0.24 |
|
|
|
| chr19_+_43524098 | 0.23 |
ENSDART00000129362
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr16_-_5205633 | 0.23 |
ENSDART00000082071
|
bckdhb
|
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr3_-_3589844 | 0.23 |
ENSDART00000140482
|
ENSDARG00000079723
|
ENSDARG00000079723 |
| chr19_-_44064157 | 0.23 |
ENSDART00000144058
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr4_+_71965053 | 0.23 |
ENSDART00000150765
|
si:ch211-165i18.2
|
si:ch211-165i18.2 |
| chr4_-_47142181 | 0.22 |
ENSDART00000150426
|
znf1064
|
zinc finger protein 1064 |
| chr1_+_53384116 | 0.22 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr11_-_232269 | 0.21 |
ENSDART00000172818
|
si:ch1073-456m8.1
|
si:ch1073-456m8.1 |
| chr4_+_13430131 | 0.21 |
ENSDART00000137200
|
si:dkey-39a18.1
|
si:dkey-39a18.1 |
| KN150266v1_+_74105 | 0.21 |
|
|
|
| chr3_-_7275872 | 0.21 |
ENSDART00000132197
|
BX005085.4
|
ENSDARG00000102658 |
| chr22_+_5633739 | 0.21 |
ENSDART00000106141
|
tma7
|
translation machinery associated 7 homolog |
| chr2_-_31627340 | 0.20 |
|
|
|
| chr14_-_16171165 | 0.20 |
ENSDART00000089021
|
canx
|
calnexin |
| chr6_-_23830912 | 0.20 |
|
|
|
| chr3_-_41649319 | 0.20 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr8_+_30912152 | 0.19 |
ENSDART00000171400
|
derl3
|
derlin 3 |
| chr10_+_26149600 | 0.17 |
|
|
|
| chr15_-_39783178 | 0.17 |
|
|
|
| chr12_-_3672263 | 0.17 |
|
|
|
| chr16_-_42857240 | 0.17 |
|
|
|
| chr9_+_37510430 | 0.17 |
ENSDART00000100622
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr10_+_21657291 | 0.16 |
|
|
|
| chr9_+_17855021 | 0.15 |
ENSDART00000166566
ENSDART00000060356 |
dgkh
|
diacylglycerol kinase, eta |
| chr14_+_11457256 | 0.14 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr22_-_19527759 | 0.14 |
ENSDART00000105485
|
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr3_+_19582238 | 0.14 |
ENSDART00000109409
|
mrc2
|
mannose receptor, C type 2 |
| chr3_+_13040717 | 0.14 |
|
|
|
| chr9_+_37510380 | 0.11 |
ENSDART00000100622
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr10_+_29425917 | 0.11 |
|
|
|
| chr3_+_23523703 | 0.10 |
ENSDART00000078428
|
hoxb13a
|
homeobox B13a |
| chr14_+_11457009 | 0.10 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr12_-_13690539 | 0.09 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr7_-_54990861 | 0.09 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr17_-_53237500 | 0.08 |
|
|
|
| chr18_+_17503828 | 0.08 |
ENSDART00000161827
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr4_+_20588066 | 0.06 |
|
|
|
| chr14_-_2379419 | 0.05 |
|
|
|
| chr14_+_26461172 | 0.05 |
ENSDART00000125289
|
ahnak
|
AHNAK nucleoprotein |
| chr18_+_41242498 | 0.03 |
ENSDART00000138552
ENSDART00000145863 |
trip12
|
thyroid hormone receptor interactor 12 |
| chr23_+_25365281 | 0.02 |
ENSDART00000016248
|
pa2g4b
|
proliferation-associated 2G4, b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.5 | 1.6 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.4 | 1.7 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.3 | 1.7 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.2 | 0.7 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 1.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 1.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.2 | 1.1 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.6 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 1.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.8 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.8 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.1 | 1.0 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.5 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.1 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 1.9 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.7 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.8 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.8 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 1.2 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.3 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.9 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 1.0 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.9 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.6 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 0.4 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 1.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.1 | GO:0015835 | peptidoglycan transport(GO:0015835) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 2.5 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.3 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 1.2 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 2.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 1.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 2.1 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.3 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.4 | 1.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.2 | 1.0 | GO:0016842 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.2 | 0.8 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.2 | 1.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 1.1 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 1.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.7 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.6 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 0.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.3 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 2.7 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 1.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.4 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 1.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 4.8 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 1.2 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |