Project
DANIO-CODE
Navigation
Downloads
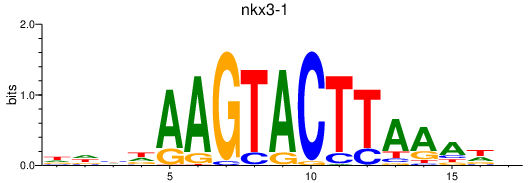
Results for nkx3-1
Z-value: 1.80
Transcription factors associated with nkx3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-1
|
ENSDARG00000078280 | NK3 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3-1 | dr10_dc_chr8_-_50270783_50270806 | -0.80 | 2.2e-04 | Click! |
Activity profile of nkx3-1 motif
Sorted Z-values of nkx3-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_34971985 | 8.12 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr9_-_8318363 | 6.00 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr7_+_17695173 | 5.67 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr16_+_54833107 | 5.10 |
ENSDART00000157641
|
fbxo43
|
F-box protein 43 |
| chr4_-_1522439 | 4.99 |
ENSDART00000175566
|
CABZ01085702.1
|
ENSDARG00000106927 |
| chr9_-_12916555 | 4.87 |
ENSDART00000140691
|
si:ch211-167j6.3
|
si:ch211-167j6.3 |
| chr22_-_12738593 | 4.49 |
ENSDART00000143574
|
rqcd1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr11_-_30388949 | 4.11 |
ENSDART00000063775
|
ENSDARG00000043442
|
ENSDARG00000043442 |
| chr10_-_34971926 | 3.93 |
ENSDART00000141201
|
ccna1
|
cyclin A1 |
| chr15_+_29092022 | 3.89 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr16_-_41585481 | 3.65 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr19_+_33552393 | 3.60 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr1_-_7160294 | 3.49 |
ENSDART00000174972
|
BX908756.2
|
ENSDARG00000108483 |
| chr19_+_33551956 | 3.43 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr5_+_36066651 | 3.40 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr22_-_7412566 | 3.25 |
ENSDART00000161615
ENSDART00000158210 |
BX511034.5
|
ENSDARG00000099295 |
| chr15_+_29092224 | 3.21 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr5_+_36066758 | 3.19 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr19_-_10411573 | 3.08 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr9_+_45193290 | 3.06 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr23_-_36178206 | 3.05 |
ENSDART00000153852
ENSDART00000153206 ENSDART00000155040 ENSDART00000156899 ENSDART00000157077 ENSDART00000155063 |
BX005254.1
|
ENSDARG00000093797 |
| chr5_-_15971338 | 2.91 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr5_-_55082078 | 2.87 |
ENSDART00000097424
|
kcmf1
|
potassium channel modulatory factor 1 |
| chr14_+_24543399 | 2.84 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr1_-_54570813 | 2.74 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr19_+_41834452 | 2.71 |
ENSDART00000148732
ENSDART00000149799 |
dlx6a
|
distal-less homeobox 6a |
| chr17_-_43040958 | 2.69 |
ENSDART00000132754
|
npc2
|
Niemann-Pick disease, type C2 |
| chr8_-_39825675 | 2.69 |
ENSDART00000130686
|
unc119.1
|
unc-119 homolog 1 |
| chr22_-_12738445 | 2.68 |
ENSDART00000047464
|
rqcd1
|
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr5_-_9577533 | 2.65 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr12_-_25058837 | 2.59 |
ENSDART00000135368
|
rhoq
|
ras homolog family member Q |
| chr5_-_55081799 | 2.59 |
ENSDART00000158102
|
kcmf1
|
potassium channel modulatory factor 1 |
| chr13_+_36554762 | 2.57 |
ENSDART00000136030
|
atp5s
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit s |
| chr14_+_50010976 | 2.55 |
ENSDART00000171955
|
CR855307.1
|
ENSDARG00000099628 |
| chr13_-_32595706 | 2.55 |
ENSDART00000012232
|
pdss2
|
prenyl (decaprenyl) diphosphate synthase, subunit 2 |
| chr5_+_36067010 | 2.47 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr24_+_25872005 | 2.43 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr19_+_2942485 | 2.42 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr11_+_36088518 | 2.42 |
|
|
|
| chr3_-_18039574 | 2.40 |
ENSDART00000049240
|
tob1a
|
transducer of ERBB2, 1a |
| chr22_-_22234380 | 2.40 |
ENSDART00000020937
|
hdgfrp2
|
hepatoma-derived growth factor-related protein 2 |
| chr10_+_33790718 | 2.38 |
ENSDART00000161430
|
rxfp2a
|
relaxin/insulin-like family peptide receptor 2a |
| chr12_-_10438255 | 2.37 |
ENSDART00000106172
|
rac1a
|
ras-related C3 botulinum toxin substrate 1a (rho family, small GTP binding protein Rac1) |
| chr12_-_31346300 | 2.32 |
ENSDART00000175929
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr9_+_28329384 | 2.31 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr2_+_10858480 | 2.30 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr17_-_15487267 | 2.27 |
|
|
|
| chr11_-_36088336 | 2.22 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr7_+_34923319 | 2.20 |
|
|
|
| chr11_-_22450582 | 2.17 |
|
|
|
| chr5_+_40543392 | 2.16 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr12_-_31346234 | 2.14 |
ENSDART00000175929
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr21_+_5027911 | 2.12 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr21_-_13654658 | 2.10 |
ENSDART00000111666
|
npdc1a
|
neural proliferation, differentiation and control, 1a |
| chr5_+_29996321 | 2.08 |
ENSDART00000124487
|
hinfp
|
histone H4 transcription factor |
| chr14_+_34673281 | 2.05 |
ENSDART00000164974
|
ebf1a
|
early B-cell factor 1a |
| chr17_+_52526741 | 2.04 |
ENSDART00000109891
|
angel1
|
angel homolog 1 (Drosophila) |
| chr16_+_16345914 | 2.03 |
ENSDART00000137672
|
setd2
|
SET domain containing 2 |
| chr17_-_43632707 | 2.01 |
ENSDART00000136431
|
rtkn2a
|
rhotekin 2a |
| chr13_-_36554590 | 2.00 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr22_+_32274436 | 1.98 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr3_+_36093650 | 1.97 |
ENSDART00000150994
|
dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr1_+_40776176 | 1.96 |
|
|
|
| chr25_-_8026912 | 1.95 |
|
|
|
| chr5_+_40543353 | 1.95 |
ENSDART00000011229
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr24_+_14451260 | 1.91 |
ENSDART00000137337
ENSDART00000091784 |
thtpa
|
thiamine triphosphatase |
| chr17_-_24684870 | 1.88 |
ENSDART00000156061
|
si:ch211-15d5.12
|
si:ch211-15d5.12 |
| chr8_-_38284959 | 1.86 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_+_18870471 | 1.86 |
ENSDART00000160185
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr5_+_56896961 | 1.84 |
|
|
|
| chr21_+_20439227 | 1.84 |
|
|
|
| chr14_-_49798900 | 1.82 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr24_+_25871962 | 1.78 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr18_+_40481566 | 1.77 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr1_+_40428827 | 1.76 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr5_+_32215942 | 1.75 |
ENSDART00000047377
|
crata
|
carnitine O-acetyltransferase a |
| chr12_-_7551118 | 1.75 |
ENSDART00000152556
|
FAM13C
|
family with sequence similarity 13 member C |
| chr20_-_13729816 | 1.74 |
ENSDART00000078893
|
ezrb
|
ezrin b |
| chr2_-_7959150 | 1.69 |
ENSDART00000136074
|
b3gnt5b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5b |
| chr5_+_17508128 | 1.66 |
ENSDART00000048859
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr25_-_13307171 | 1.66 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr10_+_44456550 | 1.65 |
ENSDART00000157611
|
snrnp35
|
small nuclear ribonucleoprotein 35 (U11/U12) |
| chr8_-_39825792 | 1.63 |
ENSDART00000130686
|
unc119.1
|
unc-119 homolog 1 |
| chr21_-_23873574 | 1.61 |
|
|
|
| chr19_-_10411518 | 1.56 |
ENSDART00000171232
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr9_-_46614763 | 1.56 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr15_+_24628191 | 1.55 |
ENSDART00000140658
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr17_-_10657734 | 1.54 |
ENSDART00000153913
|
fbxo34
|
F-box protein 34 |
| chr7_+_10669695 | 1.52 |
ENSDART00000170655
|
abhd17c
|
abhydrolase domain containing 17C |
| chr15_-_24741104 | 1.50 |
ENSDART00000100756
|
tmem199
|
transmembrane protein 199 |
| chr16_-_41585555 | 1.49 |
ENSDART00000131706
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr17_-_43041254 | 1.49 |
ENSDART00000124663
|
npc2
|
Niemann-Pick disease, type C2 |
| chr5_+_1461715 | 1.49 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr5_+_62041556 | 1.48 |
ENSDART00000146879
|
zswim7
|
zinc finger, SWIM-type containing 7 |
| chr3_-_23382550 | 1.47 |
ENSDART00000170200
|
BX682558.1
|
ENSDARG00000101975 |
| chr3_+_25719002 | 1.47 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr20_-_8847351 | 1.46 |
ENSDART00000141375
|
CR388209.1
|
ENSDARG00000091901 |
| chr7_-_48394268 | 1.45 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr21_-_14719281 | 1.44 |
ENSDART00000067004
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr16_+_43464824 | 1.43 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr3_+_16772351 | 1.43 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr7_-_60969349 | 1.42 |
ENSDART00000098610
|
rell1
|
RELT like 1 |
| chr21_+_37405864 | 1.39 |
ENSDART00000148956
|
znf346
|
zinc finger protein 346 |
| chr8_-_38284904 | 1.38 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_+_24576319 | 1.38 |
ENSDART00000164256
|
ENSDARG00000100186
|
ENSDARG00000100186 |
| chr15_+_25516764 | 1.37 |
ENSDART00000009545
|
pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr11_-_18112090 | 1.35 |
ENSDART00000019248
|
tmem110
|
transmembrane protein 110 |
| chr6_-_46733412 | 1.33 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr5_+_36066523 | 1.32 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr17_+_25407766 | 1.31 |
ENSDART00000121848
|
srrm1
|
serine/arginine repetitive matrix 1 |
| chr7_-_60969582 | 1.31 |
ENSDART00000098610
|
rell1
|
RELT like 1 |
| chr21_+_19511167 | 1.28 |
ENSDART00000058487
|
rai14
|
retinoic acid induced 14 |
| chr1_-_50626230 | 1.27 |
|
|
|
| chr8_-_31044580 | 1.26 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr11_+_26138316 | 1.25 |
ENSDART00000175997
|
cpne1
|
copine I |
| chr3_-_20807407 | 1.24 |
ENSDART00000164601
|
ndufa4l
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4, like |
| chr22_-_10511906 | 1.24 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr17_-_18878230 | 1.23 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr24_-_32607801 | 1.22 |
ENSDART00000143781
|
EIF1B
|
eukaryotic translation initiation factor 1B |
| chr20_-_26003133 | 1.19 |
ENSDART00000142611
ENSDART00000024821 |
elmsan1a
|
ELM2 and Myb/SANT-like domain containing 1a |
| chr7_-_60969664 | 1.17 |
ENSDART00000098610
|
rell1
|
RELT like 1 |
| chr7_-_26260802 | 1.16 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr19_-_18689437 | 1.16 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr6_-_46733353 | 1.15 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr6_-_29314529 | 1.15 |
ENSDART00000132456
|
bivm
|
basic, immunoglobulin-like variable motif containing |
| chr11_+_30389309 | 1.14 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr11_-_23460219 | 1.13 |
|
|
|
| chr3_-_35734943 | 1.11 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr24_-_6649190 | 1.11 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr25_+_8585585 | 1.10 |
ENSDART00000154680
|
man2a2
|
mannosidase, alpha, class 2A, member 2 |
| chr15_-_4424628 | 1.09 |
ENSDART00000062874
|
atp1b3b
|
ATPase, Na+/K+ transporting, beta 3b polypeptide |
| chr7_+_71766373 | 1.08 |
ENSDART00000011303
|
ywhae2
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide 2 |
| chr10_+_33758581 | 1.07 |
ENSDART00000141650
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr17_+_43633099 | 1.06 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr7_-_20212157 | 1.05 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr1_+_5437488 | 1.04 |
ENSDART00000092324
|
abcb6a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6a |
| chr9_-_34459768 | 1.03 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr24_-_6649248 | 1.02 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr9_-_53058151 | 1.02 |
ENSDART00000172534
ENSDART00000167922 ENSDART00000164924 |
xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr16_+_51328478 | 1.02 |
ENSDART00000168162
|
FQ323156.1
|
ENSDARG00000100697 |
| chr1_-_358810 | 1.01 |
ENSDART00000098627
|
pros1
|
protein S (alpha) |
| chr3_+_32519965 | 0.99 |
ENSDART00000131850
ENSDART00000055338 |
thoc6
|
THO complex 6 |
| chr6_-_49802414 | 0.98 |
ENSDART00000008959
|
nelfcd
|
negative elongation factor complex member C/D |
| chr6_+_19933322 | 0.98 |
ENSDART00000143378
|
thumpd3
|
THUMP domain containing 3 |
| chr24_+_15995931 | 0.97 |
ENSDART00000105955
|
TIMM21
|
translocase of inner mitochondrial membrane 21 |
| chr19_-_10411878 | 0.97 |
ENSDART00000136653
|
ccdc106b
|
coiled-coil domain containing 106b |
| chr9_+_12917201 | 0.97 |
ENSDART00000102386
|
si:ch211-167j6.4
|
si:ch211-167j6.4 |
| chr7_-_26355894 | 0.96 |
ENSDART00000159826
|
phf23b
|
PHD finger protein 23b |
| chr3_-_35735085 | 0.95 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr24_+_25872150 | 0.94 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr21_-_14719149 | 0.94 |
ENSDART00000067004
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr11_+_30389137 | 0.94 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr3_-_26675055 | 0.93 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr5_+_43455864 | 0.92 |
|
|
|
| chr7_+_30454112 | 0.92 |
ENSDART00000085716
|
mtmr10
|
myotubularin related protein 10 |
| chr1_+_40776542 | 0.91 |
ENSDART00000142967
|
dok1a
|
docking protein 1a |
| chr5_+_24378828 | 0.91 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr9_+_2472639 | 0.90 |
ENSDART00000147034
|
gpr155a
|
G protein-coupled receptor 155a |
| chr11_-_7146819 | 0.88 |
ENSDART00000172879
|
smim7
|
small integral membrane protein 7 |
| chr14_+_533611 | 0.87 |
|
|
|
| KN149789v1_-_7443 | 0.86 |
|
|
|
| chr17_+_43633129 | 0.86 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr11_-_25019165 | 0.85 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr20_-_13244237 | 0.85 |
ENSDART00000124470
|
ints7
|
integrator complex subunit 7 |
| chr20_+_31375136 | 0.85 |
|
|
|
| chr19_+_7505348 | 0.83 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr12_+_31345948 | 0.83 |
ENSDART00000031154
|
gucy2g
|
guanylate cyclase 2g |
| chr15_+_17912538 | 0.81 |
ENSDART00000168940
|
zgc:113279
|
zgc:113279 |
| chr18_-_15187432 | 0.81 |
ENSDART00000111333
|
mterf2
|
mitochondrial transcription termination factor 2 |
| chr20_+_31375056 | 0.81 |
|
|
|
| chr24_+_15996046 | 0.80 |
ENSDART00000105955
|
TIMM21
|
translocase of inner mitochondrial membrane 21 |
| chr11_-_22450525 | 0.79 |
|
|
|
| chr1_-_2309740 | 0.78 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr24_-_8217219 | 0.76 |
ENSDART00000106388
|
eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr5_+_56896748 | 0.76 |
|
|
|
| chr1_+_40776703 | 0.76 |
ENSDART00000142967
|
dok1a
|
docking protein 1a |
| chr17_-_49197264 | 0.74 |
ENSDART00000169207
|
tfb1m
|
transcription factor B1, mitochondrial |
| chr4_-_7664508 | 0.74 |
ENSDART00000148973
|
lta4h
|
leukotriene A4 hydrolase |
| chr5_-_68935171 | 0.74 |
ENSDART00000149692
|
dguok
|
deoxyguanosine kinase |
| chr22_+_520271 | 0.74 |
ENSDART00000067638
|
med20
|
mediator complex subunit 20 |
| chr2_-_37555425 | 0.73 |
ENSDART00000143496
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr18_-_48329076 | 0.72 |
|
|
|
| chr14_+_51724531 | 0.72 |
ENSDART00000168437
|
b4galt1l
|
DP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1, like |
| chr17_+_13654337 | 0.72 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr3_-_36277758 | 0.70 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr21_+_1719308 | 0.70 |
|
|
|
| chr2_-_56007484 | 0.70 |
ENSDART00000097755
|
jund
|
jun D proto-oncogene |
| chr23_+_7614867 | 0.69 |
ENSDART00000011554
|
tm9sf4
|
transmembrane 9 superfamily protein member 4 |
| chr8_+_42968542 | 0.68 |
|
|
|
| chr20_+_25652930 | 0.67 |
ENSDART00000162080
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr19_+_46371192 | 0.67 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr7_+_22386328 | 0.66 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr3_+_25718973 | 0.66 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr21_-_35291005 | 0.65 |
ENSDART00000134780
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr17_-_24684729 | 0.65 |
|
|
|
| chr7_+_57877047 | 0.65 |
ENSDART00000123117
|
ggh
|
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
| chr9_-_11683921 | 0.64 |
ENSDART00000044314
ENSDART00000124129 |
itgav
|
integrin, alpha V |
| chr23_-_15090815 | 0.64 |
ENSDART00000147636
|
ndrg3b
|
ndrg family member 3b |
| chr8_-_6782167 | 0.62 |
ENSDART00000135834
ENSDART00000172157 |
dock5
|
dedicator of cytokinesis 5 |
| chr19_+_7839888 | 0.62 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:0032890 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 1.3 | 3.9 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 1.1 | 5.6 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 1.0 | 2.9 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.9 | 4.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.8 | 2.5 | GO:0070922 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) small RNA loading onto RISC(GO:0070922) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.6 | 5.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.6 | 1.9 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.6 | 5.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of meiotic cell cycle(GO:0051447) |
| 0.6 | 1.9 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.6 | 2.4 | GO:0030801 | positive regulation of cyclic nucleotide metabolic process(GO:0030801) positive regulation of cyclic nucleotide biosynthetic process(GO:0030804) positive regulation of nucleotide biosynthetic process(GO:0030810) positive regulation of cyclase activity(GO:0031281) positive regulation of lyase activity(GO:0051349) positive regulation of purine nucleotide biosynthetic process(GO:1900373) |
| 0.5 | 1.5 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.4 | 1.8 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.4 | 1.8 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.3 | GO:1903894 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.4 | 1.2 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.4 | 1.2 | GO:0019376 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.4 | 4.1 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.3 | 1.0 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.3 | 1.9 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.3 | 4.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.3 | 1.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 0.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.9 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.2 | 1.7 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 0.8 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.2 | 11.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.2 | 2.7 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 1.4 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.2 | 1.0 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.8 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.2 | 1.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.7 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 2.6 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.1 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 2.7 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.1 | 2.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) regulation of neutrophil migration(GO:1902622) |
| 0.1 | 1.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 2.0 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.7 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 2.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 2.6 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 2.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 0.3 | GO:0014807 | regulation of somitogenesis(GO:0014807) regulation of heart looping(GO:1901207) positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 1.4 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 1.1 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 1.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.8 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.9 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 2.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 5.2 | GO:0099536 | synaptic signaling(GO:0099536) |
| 0.0 | 0.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.9 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 9.3 | GO:0006915 | apoptotic process(GO:0006915) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 1.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 3.0 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.5 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.7 | 5.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.5 | 7.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 1.5 | GO:0097196 | Shu complex(GO:0097196) |
| 0.4 | 2.9 | GO:0071546 | pi-body(GO:0071546) |
| 0.4 | 2.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 1.7 | GO:0000811 | GINS complex(GO:0000811) |
| 0.2 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 2.5 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.2 | 2.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 2.7 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 5.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 2.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 2.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.1 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 1.5 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.0 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 3.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 2.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 4.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.9 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.7 | 2.9 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.6 | 2.4 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.6 | 1.8 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.5 | 1.9 | GO:0003823 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.4 | 4.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.4 | 1.2 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.4 | 2.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.4 | 1.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 2.5 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 1.8 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.3 | 1.4 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.3 | 0.8 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.7 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.2 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 11.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 1.9 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 2.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 1.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.4 | GO:0001653 | peptide receptor activity(GO:0001653) G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 2.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 2.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 2.1 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.0 | 1.7 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 5.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 2.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 4.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 2.0 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 1.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.9 | GO:0032182 | ubiquitin-like protein binding(GO:0032182) |
| 0.0 | 5.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 2.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 12.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 1.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 12.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.6 | 4.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.4 | 2.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.4 | 7.0 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 1.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.3 | 1.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 2.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 2.0 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |