Project
DANIO-CODE
Navigation
Downloads
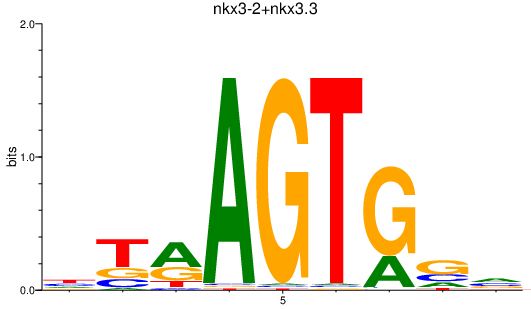
Results for nkx3-2+nkx3.3
Z-value: 0.69
Transcription factors associated with nkx3-2+nkx3.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx3-2
|
ENSDARG00000037639 | NK3 homeobox 2 |
|
nkx3.3
|
ENSDARG00000069327 | NK3 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx3.2 | dr10_dc_chr14_-_7772_7799 | -0.40 | 1.2e-01 | Click! |
Activity profile of nkx3-2+nkx3.3 motif
Sorted Z-values of nkx3-2+nkx3.3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx3-2+nkx3.3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| KN150589v1_-_5209 | 2.17 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr13_+_8508786 | 2.10 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr19_+_40558066 | 1.41 |
ENSDART00000049968
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr1_-_51075777 | 1.33 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr24_+_21531899 | 1.15 |
ENSDART00000158833
|
si:ch211-140b10.6
|
si:ch211-140b10.6 |
| chr25_+_19572283 | 1.14 |
ENSDART00000073472
|
zgc:113426
|
zgc:113426 |
| chr8_-_3974320 | 1.13 |
ENSDART00000159895
|
mtmr3
|
myotubularin related protein 3 |
| chr24_+_16861121 | 1.05 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr5_-_26164683 | 1.00 |
ENSDART00000140392
|
rnf181
|
ring finger protein 181 |
| chr15_+_24741931 | 0.98 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr11_-_24300905 | 0.92 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr8_-_37023410 | 0.92 |
ENSDART00000111513
|
si:ch211-218o21.4
|
si:ch211-218o21.4 |
| chr19_+_20404995 | 0.89 |
ENSDART00000142841
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr15_+_24741620 | 0.88 |
ENSDART00000078014
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr21_+_20350152 | 0.87 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr21_-_23873574 | 0.86 |
|
|
|
| chr3_+_62051674 | 0.86 |
ENSDART00000113144
|
zgc:173575
|
zgc:173575 |
| chr16_-_41718971 | 0.81 |
|
|
|
| chr5_-_26164728 | 0.78 |
ENSDART00000140392
|
rnf181
|
ring finger protein 181 |
| chr19_+_20405107 | 0.74 |
ENSDART00000151066
|
osbpl3a
|
oxysterol binding protein-like 3a |
| chr16_+_25248840 | 0.73 |
ENSDART00000179520
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr11_-_24300849 | 0.67 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr7_+_53952626 | 0.66 |
ENSDART00000158518
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_30388949 | 0.64 |
ENSDART00000063775
|
ENSDARG00000043442
|
ENSDARG00000043442 |
| chr21_-_2322963 | 0.61 |
ENSDART00000160337
|
si:ch73-299h12.8
|
si:ch73-299h12.8 |
| chr20_+_49380442 | 0.61 |
|
|
|
| chr7_+_13573107 | 0.60 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr25_-_19927083 | 0.59 |
ENSDART00000140182
|
cnot4a
|
CCR4-NOT transcription complex, subunit 4a |
| chr6_-_21755349 | 0.58 |
|
|
|
| chr25_+_16259634 | 0.58 |
ENSDART00000136454
|
tead1a
|
TEA domain family member 1a |
| chr10_-_29945491 | 0.57 |
ENSDART00000063923
ENSDART00000136264 |
zpr1
|
ZPR1 zinc finger |
| chr16_+_25248941 | 0.55 |
ENSDART00000179520
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr20_+_6783996 | 0.55 |
ENSDART00000169966
|
igfbp3
|
insulin-like growth factor binding protein 3 |
| chr6_-_21756853 | 0.55 |
ENSDART00000169035
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr5_-_22090338 | 0.53 |
|
|
|
| chr20_+_30475949 | 0.51 |
ENSDART00000148242
|
rnaseh1
|
ribonuclease H1 |
| chr1_+_15928362 | 0.51 |
ENSDART00000149026
ENSDART00000159785 ENSDART00000164899 |
pcm1
|
pericentriolar material 1 |
| chr11_+_30389309 | 0.50 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr7_+_53952463 | 0.48 |
ENSDART00000158518
|
pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr13_-_24614604 | 0.48 |
ENSDART00000031564
|
sfr1
|
SWI5-dependent homologous recombination repair protein 1 |
| chr20_-_23595575 | 0.46 |
ENSDART00000174638
|
anapc4
|
anaphase promoting complex subunit 4 |
| chr11_-_24300785 | 0.45 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr18_-_20469413 | 0.45 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr18_+_22185449 | 0.43 |
ENSDART00000089549
|
fam65a
|
family with sequence similarity 65, member A |
| chr18_-_20469253 | 0.43 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr5_-_22090397 | 0.43 |
|
|
|
| chr12_-_13690748 | 0.42 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr1_+_28860492 | 0.42 |
ENSDART00000154342
ENSDART00000088290 |
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr8_+_25279900 | 0.41 |
ENSDART00000114676
|
gstm.2
|
glutathione S-transferase mu tandem duplicate 2 |
| chr17_+_24699773 | 0.40 |
ENSDART00000007271
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr4_+_13932693 | 0.39 |
ENSDART00000142466
|
pphln1
|
periphilin 1 |
| chr18_+_22185526 | 0.39 |
ENSDART00000089549
|
fam65a
|
family with sequence similarity 65, member A |
| chr20_-_8847351 | 0.39 |
ENSDART00000141375
|
CR388209.1
|
ENSDARG00000091901 |
| chr15_-_24007305 | 0.38 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr22_+_2386743 | 0.38 |
ENSDART00000132925
|
zgc:112977
|
zgc:112977 |
| chr2_-_40067342 | 0.38 |
|
|
|
| chr7_-_40307611 | 0.35 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr11_-_24301030 | 0.34 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr3_+_52951742 | 0.33 |
|
|
|
| chr11_+_30389137 | 0.33 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr10_+_24079292 | 0.32 |
ENSDART00000149265
|
gbe1a
|
glucan (1,4-alpha-), branching enzyme 1a |
| chr8_-_48858897 | 0.32 |
ENSDART00000122458
ENSDART00000010591 |
wrap73
|
WD repeat containing, antisense to TP73 |
| chr11_-_5956333 | 0.31 |
ENSDART00000123601
|
dda1
|
DET1 and DDB1 associated 1 |
| chr15_-_24007488 | 0.31 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr12_-_13690539 | 0.30 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr16_-_29470491 | 0.30 |
ENSDART00000175571
|
tlr18
|
toll-like receptor 18 |
| chr15_+_24741845 | 0.27 |
ENSDART00000143137
|
poldip2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr4_-_11126782 | 0.25 |
ENSDART00000150419
|
BX784029.1
|
ENSDARG00000096039 |
| chr18_-_22105000 | 0.25 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr15_-_24007354 | 0.25 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr16_-_22669752 | 0.24 |
ENSDART00000134239
|
si:dkey-238m4.3
|
si:dkey-238m4.3 |
| chr14_-_1186603 | 0.22 |
|
|
|
| chr18_-_22185401 | 0.21 |
|
|
|
| chr7_-_28300008 | 0.20 |
|
|
|
| chr11_+_6450876 | 0.20 |
ENSDART00000170180
|
CU062501.2
|
ENSDARG00000103967 |
| chr5_+_26165043 | 0.20 |
ENSDART00000144169
|
si:ch211-102c2.8
|
si:ch211-102c2.8 |
| chr7_+_71255442 | 0.19 |
|
|
|
| chr11_-_25495956 | 0.19 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr12_+_22459371 | 0.16 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr3_-_36126124 | 0.16 |
|
|
|
| chr15_-_38027496 | 0.16 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr21_-_1956120 | 0.15 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr7_+_13573147 | 0.13 |
ENSDART00000035067
|
abhd2a
|
abhydrolase domain containing 2a |
| chr15_-_38027363 | 0.12 |
ENSDART00000157550
|
si:dkey-238d18.15
|
si:dkey-238d18.15 |
| chr6_-_39201256 | 0.12 |
ENSDART00000077938
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr13_+_24453943 | 0.09 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr24_-_31285903 | 0.09 |
ENSDART00000164001
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr21_+_1997858 | 0.07 |
|
|
|
| chr5_-_48377307 | 0.07 |
ENSDART00000097452
|
si:dkey-172m14.1
|
si:dkey-172m14.1 |
| chr25_+_35762694 | 0.06 |
ENSDART00000136456
ENSDART00000175090 |
CR354435.8
zgc:173552
|
Histone H3.2 zgc:173552 |
| chr14_-_1186789 | 0.06 |
|
|
|
| chr14_-_1186858 | 0.04 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr5_-_22613185 | 0.03 |
ENSDART00000051529
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr15_+_862607 | 0.03 |
ENSDART00000174813
|
si:dkey-7i4.7
|
si:dkey-7i4.7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.7 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.7 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.4 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 1.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 2.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 2.1 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 2.1 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 0.5 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.4 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.3 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 2.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 1.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.5 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.1 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 2.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0034399 | anaphase-promoting complex(GO:0005680) nuclear periphery(GO:0034399) |
| 0.0 | 0.9 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.1 | 1.3 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.2 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 2.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 1.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |