Project
DANIO-CODE
Navigation
Downloads
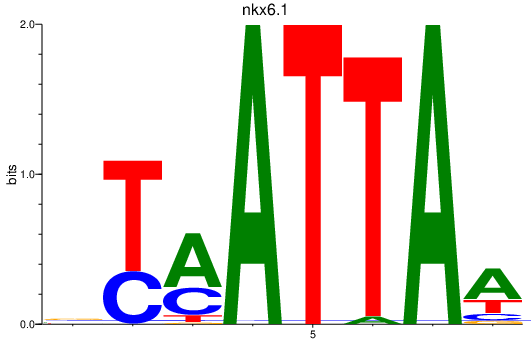
Results for nkx6.1
Z-value: 1.00
Transcription factors associated with nkx6.1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.1
|
ENSDARG00000022569 | NK6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nkx6.1 | dr10_dc_chr21_+_19034242_19034288 | 0.88 | 7.8e-06 | Click! |
Activity profile of nkx6.1 motif
Sorted Z-values of nkx6.1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_22576513 | 3.82 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr16_+_46145286 | 3.73 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr12_+_24221087 | 2.94 |
ENSDART00000088178
|
nrxn1a
|
neurexin 1a |
| chr9_-_32942783 | 2.83 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr10_+_39141022 | 2.41 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr16_+_23516127 | 2.35 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr14_+_46419051 | 2.25 |
ENSDART00000112377
|
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr6_+_56157608 | 2.10 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr17_+_23278879 | 2.08 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr24_-_21778717 | 2.03 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr2_+_49358871 | 1.96 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr3_+_56970554 | 1.95 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr20_-_29517770 | 1.94 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr7_-_25623974 | 1.93 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr23_-_7740845 | 1.93 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr17_+_15425559 | 1.89 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr15_-_4537178 | 1.88 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr12_+_20230575 | 1.81 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr11_+_23522743 | 1.79 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr15_-_18159904 | 1.75 |
ENSDART00000170874
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr21_+_28441951 | 1.75 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr18_-_3056732 | 1.71 |
ENSDART00000162657
|
rps3
|
ribosomal protein S3 |
| chr17_-_29102320 | 1.70 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr20_-_9107294 | 1.68 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr11_+_10925475 | 1.68 |
ENSDART00000064860
|
rbms1a
|
RNA binding motif, single stranded interacting protein 1a |
| chr24_+_5205878 | 1.63 |
ENSDART00000106488
ENSDART00000005901 |
plod2
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr14_+_20045365 | 1.58 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr18_+_26437604 | 1.56 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr5_+_38236956 | 1.56 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr11_+_14142126 | 1.55 |
ENSDART00000102520
|
palm1a
|
paralemmin 1a |
| chr1_-_4757890 | 1.50 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr4_-_15442828 | 1.50 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr6_-_54816183 | 1.47 |
ENSDART00000148462
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr6_+_3873114 | 1.47 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr16_-_42990753 | 1.47 |
ENSDART00000149317
|
hfe2
|
hemochromatosis type 2 |
| chr20_+_33391554 | 1.46 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr22_-_37417903 | 1.44 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr1_+_13779755 | 1.44 |
|
|
|
| chr18_+_29424354 | 1.43 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr16_-_17439735 | 1.43 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr3_-_35928594 | 1.38 |
|
|
|
| chr20_-_3970778 | 1.38 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr7_+_6517248 | 1.38 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr21_+_28408329 | 1.37 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr19_+_32579358 | 1.37 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr21_-_22510751 | 1.37 |
ENSDART00000169870
|
myo5b
|
myosin VB |
| chr16_+_23172295 | 1.36 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr24_-_6048914 | 1.36 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr25_+_14411153 | 1.35 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr8_+_39964695 | 1.34 |
ENSDART00000073782
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr3_+_17387551 | 1.33 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr5_-_40707316 | 1.33 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr14_-_17258072 | 1.33 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr23_-_23474703 | 1.32 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr18_-_43890514 | 1.31 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr1_+_26330186 | 1.28 |
ENSDART00000102322
|
bnc2
|
basonuclin 2 |
| chr2_-_34010299 | 1.28 |
ENSDART00000140910
|
ptch2
|
patched 2 |
| chr3_-_6078015 | 1.28 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr3_-_29768364 | 1.27 |
ENSDART00000020311
|
rpl27
|
ribosomal protein L27 |
| chr16_+_13928844 | 1.24 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr22_-_13018196 | 1.24 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr21_-_25567978 | 1.24 |
ENSDART00000133134
|
efemp2b
|
EGF containing fibulin-like extracellular matrix protein 2b |
| chr1_-_40208544 | 1.23 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr23_+_36023748 | 1.21 |
|
|
|
| chr14_-_17257773 | 1.21 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr6_+_23787804 | 1.21 |
ENSDART00000163188
|
znf648
|
zinc finger protein 648 |
| chr24_+_21395671 | 1.21 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr16_-_28943421 | 1.20 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr7_+_34018862 | 1.19 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr21_+_17731439 | 1.19 |
ENSDART00000124173
|
rxraa
|
retinoid X receptor, alpha a |
| chr16_+_29060022 | 1.18 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr3_-_45779817 | 1.16 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr23_+_20936374 | 1.16 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr23_+_11350727 | 1.14 |
|
|
|
| chr22_+_16509286 | 1.14 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr12_-_10372055 | 1.13 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr18_+_48426375 | 1.13 |
|
|
|
| chr4_-_2615160 | 1.12 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr21_-_11539798 | 1.12 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr1_-_40208469 | 1.12 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr2_-_23348612 | 1.12 |
ENSDART00000110373
|
znf414
|
zinc finger protein 414 |
| chr2_-_10555152 | 1.12 |
ENSDART00000150166
|
gng12a
|
guanine nucleotide binding protein (G protein), gamma 12a |
| chr2_-_36058327 | 1.11 |
ENSDART00000003550
|
nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr9_-_20562293 | 1.11 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr19_-_7069920 | 1.10 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr18_-_19467100 | 1.10 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr9_-_31467299 | 1.10 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr24_-_14447773 | 1.09 |
|
|
|
| chr5_-_71643185 | 1.08 |
ENSDART00000029014
|
pax8
|
paired box 8 |
| chr19_+_31046291 | 1.08 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr20_-_22478716 | 1.08 |
ENSDART00000110967
ENSDART00000011135 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr15_-_20088439 | 1.07 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr22_+_12746080 | 1.06 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr8_-_34077387 | 1.06 |
ENSDART00000159208
ENSDART00000040126 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr14_-_4166292 | 1.05 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr14_-_20760787 | 1.05 |
|
|
|
| chr9_-_43736549 | 1.05 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr16_+_26900732 | 1.04 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr2_-_40170303 | 1.04 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr21_+_28921734 | 1.04 |
ENSDART00000166575
|
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_-_5693715 | 1.03 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr10_-_33678206 | 1.02 |
ENSDART00000128049
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr14_+_14356509 | 1.01 |
ENSDART00000170211
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr24_+_17125429 | 1.01 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr15_-_14616083 | 1.01 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr6_-_2294751 | 1.01 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr7_+_19300351 | 1.00 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr6_-_43094573 | 1.00 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr25_+_2933235 | 0.99 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr1_-_518543 | 0.98 |
ENSDART00000138032
|
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr19_+_22478256 | 0.98 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr17_-_31642673 | 0.98 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr2_+_33907298 | 0.97 |
|
|
|
| chr8_-_23591082 | 0.97 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr5_-_67233396 | 0.96 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr13_+_28617462 | 0.96 |
ENSDART00000101633
|
prom2
|
prominin 2 |
| chr13_+_33151628 | 0.96 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr16_-_13733759 | 0.96 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr3_+_61924544 | 0.95 |
ENSDART00000090370
|
noxo1a
|
NADPH oxidase organizer 1a |
| chr25_+_24193604 | 0.95 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr20_-_21031504 | 0.95 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr22_-_13325409 | 0.95 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr2_+_33343287 | 0.94 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr16_+_24766043 | 0.94 |
ENSDART00000114304
|
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr13_-_252864 | 0.94 |
|
|
|
| chr1_-_11474337 | 0.93 |
ENSDART00000149913
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr15_-_1858350 | 0.93 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr5_-_32687349 | 0.93 |
ENSDART00000004238
|
rpl7a
|
ribosomal protein L7a |
| chr5_+_62987426 | 0.92 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr20_-_14158114 | 0.92 |
ENSDART00000009549
|
rhag
|
Rh-associated glycoprotein |
| chr11_+_36983626 | 0.90 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr6_+_24299180 | 0.90 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr20_+_34417665 | 0.89 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr19_-_7069850 | 0.89 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr1_-_30801520 | 0.89 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr21_+_10663517 | 0.88 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr24_+_30343717 | 0.88 |
ENSDART00000162377
|
FP085414.1
|
ENSDARG00000100270 |
| chr13_-_31165867 | 0.88 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr8_-_19166630 | 0.88 |
|
|
|
| chr5_+_36431824 | 0.87 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr7_-_58649991 | 0.86 |
ENSDART00000167076
|
mrc1a
|
mannose receptor, C type 1a |
| chr9_-_41608298 | 0.86 |
|
|
|
| chr23_+_23094125 | 0.86 |
ENSDART00000146415
ENSDART00000146463 |
samd11
|
sterile alpha motif domain containing 11 |
| chr1_-_25600988 | 0.86 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr10_+_38582701 | 0.85 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr15_+_29343644 | 0.85 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr18_+_24935499 | 0.85 |
|
|
|
| chr3_+_18406137 | 0.85 |
ENSDART00000158791
|
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr17_+_30352361 | 0.85 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr14_+_30568242 | 0.85 |
|
|
|
| chr19_-_31815128 | 0.84 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr23_+_35996491 | 0.84 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr6_-_8075384 | 0.84 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr14_-_2682064 | 0.83 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr3_-_34208423 | 0.83 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr17_+_25500291 | 0.83 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr20_+_28531684 | 0.83 |
ENSDART00000123387
|
dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr15_+_46141934 | 0.83 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr23_+_6043862 | 0.82 |
|
|
|
| chr2_+_3370130 | 0.82 |
ENSDART00000098391
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr15_-_23441268 | 0.82 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr25_-_5883620 | 0.82 |
ENSDART00000136054
|
snx22
|
sorting nexin 22 |
| chr22_-_15930756 | 0.82 |
ENSDART00000080047
|
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr3_-_41370116 | 0.82 |
|
|
|
| chr14_+_2969679 | 0.81 |
ENSDART00000044678
|
ENSDARG00000034011
|
ENSDARG00000034011 |
| chr12_-_26339002 | 0.81 |
ENSDART00000153214
|
synpo2lb
|
synaptopodin 2-like b |
| chr11_-_18068313 | 0.79 |
|
|
|
| chr11_-_37730181 | 0.79 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr17_-_47893203 | 0.78 |
|
|
|
| chr22_-_864745 | 0.78 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr8_+_26502828 | 0.76 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr5_-_33156615 | 0.76 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr23_-_22186674 | 0.76 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr3_-_39554155 | 0.75 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr17_+_29259577 | 0.75 |
ENSDART00000049399
|
ankrd9
|
ankyrin repeat domain 9 |
| chr11_+_14373731 | 0.74 |
ENSDART00000161879
|
BX571942.1
|
ENSDARG00000099220 |
| chr14_+_36156947 | 0.74 |
|
|
|
| chr10_-_43200499 | 0.74 |
ENSDART00000171494
|
ssbp2
|
single-stranded DNA binding protein 2 |
| chr5_-_44229464 | 0.74 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr16_-_45211650 | 0.73 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr13_-_28177612 | 0.73 |
ENSDART00000045351
|
lbx1a
|
ladybird homeobox 1a |
| chr10_+_13042496 | 0.73 |
ENSDART00000158919
|
lpar1
|
lysophosphatidic acid receptor 1 |
| chr25_+_28717079 | 0.73 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase, muscle, b |
| chr8_-_11512545 | 0.72 |
ENSDART00000133932
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr7_+_54410044 | 0.72 |
|
|
|
| chr16_-_41515399 | 0.71 |
ENSDART00000169116
|
cpne4a
|
copine IVa |
| chr20_-_37587804 | 0.71 |
ENSDART00000166106
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr10_+_32739166 | 0.71 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr24_+_9272045 | 0.70 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr21_+_6858073 | 0.69 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr19_+_5562107 | 0.68 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr5_-_47520995 | 0.68 |
ENSDART00000144252
|
BX465834.1
|
ENSDARG00000095715 |
| chr18_-_7184699 | 0.68 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr14_+_160149 | 0.68 |
ENSDART00000158405
ENSDART00000158072 |
gpc2
|
glypican 2 |
| chr25_+_2909562 | 0.68 |
ENSDART00000149360
|
mpi
|
mannose phosphate isomerase |
| chr9_+_34832049 | 0.68 |
ENSDART00000100735
ENSDART00000133996 |
shox
|
short stature homeobox |
| chr8_-_17580655 | 0.67 |
|
|
|
| chr20_-_23526954 | 0.67 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr9_-_3700395 | 0.67 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr3_+_28822408 | 0.66 |
ENSDART00000133528
|
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.5 | 1.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.5 | 1.4 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 1.3 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.4 | 3.8 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.4 | 1.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.4 | 1.4 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 0.7 | GO:0050886 | endocrine process(GO:0050886) |
| 0.4 | 1.1 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.3 | 0.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 0.9 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 1.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.3 | 2.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.3 | 1.0 | GO:0021559 | auditory receptor cell fate commitment(GO:0009912) trigeminal nerve development(GO:0021559) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.3 | 0.8 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 2.5 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 1.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.2 | 0.9 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.2 | 1.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.7 | GO:0090171 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) chondrocyte morphogenesis(GO:0090171) |
| 0.2 | 1.6 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 0.6 | GO:0014005 | regulation of cellular amino acid metabolic process(GO:0006521) microglia development(GO:0014005) regulation of cellular amine metabolic process(GO:0033238) |
| 0.2 | 1.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.2 | 0.8 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 2.0 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 1.8 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.2 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 0.6 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.2 | 2.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.2 | 0.5 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.2 | 1.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.2 | 1.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.0 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 2.1 | GO:0071698 | olfactory placode development(GO:0071698) |
| 0.2 | 1.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 2.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.0 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 0.4 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.4 | GO:1902871 | negative regulation of photoreceptor cell differentiation(GO:0046533) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.6 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.1 | 1.5 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.4 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.1 | GO:0034672 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 | 1.4 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.7 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 0.3 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.9 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.7 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.1 | 0.8 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 4.7 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 1.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.0 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 1.6 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.1 | 1.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.5 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 1.7 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.1 | 0.9 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 1.9 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 0.6 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.5 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.0 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 0.4 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.4 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 1.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.8 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 4.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.8 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.8 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.3 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 1.0 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 1.0 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.6 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 1.0 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 1.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.5 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 4.0 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.2 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.7 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.2 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.3 | GO:0071248 | cellular response to metal ion(GO:0071248) |
| 0.0 | 1.2 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.2 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.8 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.8 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.9 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 1.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.3 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) maintenance of protein localization in endoplasmic reticulum(GO:0035437) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.3 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 1.2 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 2.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 1.4 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.7 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.1 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 1.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0015729 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.0 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 1.1 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 1.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.3 | 1.9 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.3 | 1.0 | GO:0071914 | prominosome(GO:0071914) |
| 0.3 | 1.8 | GO:0005833 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 1.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 1.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.0 | GO:0030677 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.7 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 3.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 0.8 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.5 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.4 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 2.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 6.3 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 1.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 3.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.1 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 5.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.5 | 1.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.4 | 1.1 | GO:0005542 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
| 0.3 | 1.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 1.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.3 | 1.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 1.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.3 | 1.4 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.8 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.3 | 1.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.2 | 1.0 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.2 | 0.7 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 1.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.2 | 1.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 0.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.2 | 1.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.9 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.2 | 0.7 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.9 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.2 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 1.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.6 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 1.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 1.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.0 | GO:0004322 | ferroxidase activity(GO:0004322) ferric iron binding(GO:0008199) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.5 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 5.9 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 1.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 2.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 2.0 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 4.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.6 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 3.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.2 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 1.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.1 | 1.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.1 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.7 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0051192 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 4.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 1.7 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 33.5 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 1.7 | GO:0044389 | ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 3.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.0 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 1.0 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 1.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.2 | 1.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.2 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.9 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 3.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.4 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 0.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 5.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.5 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.1 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |