Project
DANIO-CODE
Navigation
Downloads
Results for nkx6.3
Z-value: 0.23
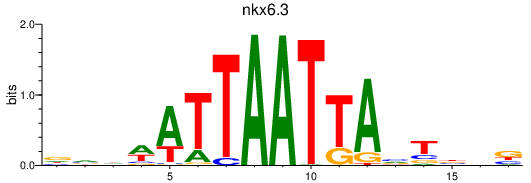
Transcription factors associated with nkx6.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nkx6.3
|
ENSDARG00000060529 | NK6 homeobox 3 |
Activity profile of nkx6.3 motif
Sorted Z-values of nkx6.3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nkx6.3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_46145286 | 1.08 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr21_+_28408329 | 0.65 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr17_+_15425559 | 0.64 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr10_-_15896595 | 0.57 |
ENSDART00000092343
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr1_+_13779755 | 0.55 |
|
|
|
| chr20_-_9107294 | 0.55 |
ENSDART00000140792
|
OMA1
|
OMA1 zinc metallopeptidase |
| chr14_+_14356509 | 0.50 |
ENSDART00000170211
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr20_-_48677794 | 0.46 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr12_-_10372055 | 0.42 |
ENSDART00000052001
|
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr5_+_62987426 | 0.42 |
ENSDART00000178937
|
dnm1b
|
dynamin 1b |
| chr6_+_3873114 | 0.42 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr6_-_16590883 | 0.38 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr25_+_24193604 | 0.33 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr17_+_25500291 | 0.31 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr16_-_12282596 | 0.29 |
ENSDART00000110567
|
clstn3
|
calsyntenin 3 |
| chr3_-_59884282 | 0.21 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr18_-_7184699 | 0.19 |
ENSDART00000135587
|
cd9a
|
CD9 molecule a |
| chr9_-_43736549 | 0.15 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr18_+_1353738 | 0.13 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr2_-_10020770 | 0.13 |
ENSDART00000046587
|
ap2m1a
|
adaptor-related protein complex 2, mu 1 subunit, a |
| chr18_+_1353857 | 0.12 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr3_-_20861178 | 0.12 |
ENSDART00000109578
|
fam117aa
|
family with sequence similarity 117, member Aa |
| chr22_-_12905648 | 0.10 |
ENSDART00000157820
|
mfsd6a
|
major facilitator superfamily domain containing 6a |
| chr3_+_18645970 | 0.09 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr2_+_1881022 | 0.09 |
ENSDART00000101038
|
tmie
|
transmembrane inner ear |
| chr9_+_31204464 | 0.08 |
|
|
|
| chr13_+_22346304 | 0.06 |
ENSDART00000137220
|
ldb3a
|
LIM domain binding 3a |
| chr23_-_35691396 | 0.05 |
ENSDART00000011004
|
mfsd5
|
major facilitator superfamily domain containing 5 |
| chr1_-_18059469 | 0.05 |
ENSDART00000140981
|
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr6_+_54179493 | 0.02 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr8_+_8889274 | 0.01 |
ENSDART00000142075
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr10_-_20545884 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.5 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.6 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0051965 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.0 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 1.1 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.3 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.1 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |