Project
DANIO-CODE
Navigation
Downloads
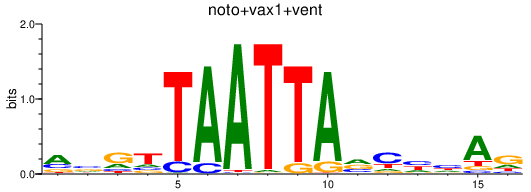
Results for noto+vax1+vent
Z-value: 0.41
Transcription factors associated with noto+vax1+vent
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
noto
|
ENSDARG00000021201 | notochord homeobox |
|
vax1
|
ENSDARG00000021916 | ventral anterior homeobox 1 |
|
vent
|
ENSDARG00000100483 | ventral expressed homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| vax1 | dr10_dc_chr17_-_21422311_21422392 | -0.57 | 2.0e-02 | Click! |
| vent | dr10_dc_chr13_-_50299821_50299866 | -0.23 | 3.9e-01 | Click! |
| noto | dr10_dc_chr13_+_14845093_14845100 | -0.20 | 4.6e-01 | Click! |
Activity profile of noto+vax1+vent motif
Sorted Z-values of noto+vax1+vent motif
Network of associatons between targets according to the STRING database.
First level regulatory network of noto+vax1+vent
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_36168475 | 0.84 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr3_+_479366 | 0.73 |
|
|
|
| chr25_+_35788501 | 0.72 |
ENSDART00000121838
|
hist1h4l
|
histone 1, H4, like |
| chr17_+_16038358 | 0.71 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr10_+_15066791 | 0.68 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr22_-_21873054 | 0.66 |
|
|
|
| chr21_+_19040595 | 0.59 |
ENSDART00000145969
|
BX000991.1
|
ENSDARG00000092282 |
| chr5_-_23211957 | 0.58 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr12_+_47448318 | 0.57 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr24_-_14446593 | 0.56 |
|
|
|
| chr15_+_35089305 | 0.53 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr21_-_3548863 | 0.48 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr10_+_15066970 | 0.46 |
ENSDART00000140084
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr18_-_43890836 | 0.46 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr10_+_15066654 | 0.46 |
ENSDART00000135667
|
si:dkey-88l16.5
|
si:dkey-88l16.5 |
| chr24_+_19270877 | 0.44 |
|
|
|
| chr20_-_37910887 | 0.41 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr9_+_30610756 | 0.40 |
ENSDART00000145025
|
zgc:113314
|
zgc:113314 |
| chr11_+_11319810 | 0.38 |
ENSDART00000162486
|
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr20_+_29306677 | 0.37 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr20_-_16553968 | 0.36 |
ENSDART00000159053
|
gtf2a1
|
general transcription factor IIA, 1 |
| chr15_-_43402935 | 0.34 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr23_+_33792145 | 0.33 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr22_-_21872864 | 0.31 |
ENSDART00000158501
|
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr12_+_22459177 | 0.31 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_+_35790782 | 0.31 |
ENSDART00000125221
|
CR933559.1
|
ENSDARG00000087939 |
| chr23_-_33692244 | 0.31 |
|
|
|
| chr12_+_34790513 | 0.30 |
ENSDART00000015643
|
tbcc
|
tubulin folding cofactor C |
| chr20_+_29306863 | 0.28 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr12_-_20494169 | 0.27 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr5_-_14783255 | 0.27 |
ENSDART00000052712
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr6_-_22799815 | 0.26 |
ENSDART00000165429
|
zgc:174863
|
zgc:174863 |
| chr3_-_27470798 | 0.26 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr7_-_58863023 | 0.25 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr6_-_30967242 | 0.24 |
ENSDART00000154523
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr20_+_29306945 | 0.23 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr6_-_40411469 | 0.23 |
ENSDART00000154520
|
BX936393.1
|
ENSDARG00000097645 |
| chr21_+_8334453 | 0.20 |
ENSDART00000055336
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr22_-_2942708 | 0.19 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr3_+_33608669 | 0.19 |
|
|
|
| chr3_+_33608622 | 0.18 |
|
|
|
| chr12_+_3632548 | 0.18 |
|
|
|
| chr7_-_58863056 | 0.17 |
ENSDART00000170853
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr6_+_40925259 | 0.15 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr2_+_50873843 | 0.13 |
|
|
|
| chr15_-_5911817 | 0.13 |
|
|
|
| chr10_-_44170848 | 0.12 |
ENSDART00000135240
|
acads
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr20_+_29307283 | 0.12 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr15_-_5912105 | 0.11 |
|
|
|
| chr19_+_21783204 | 0.11 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr8_-_25015215 | 0.10 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr16_-_7911453 | 0.10 |
ENSDART00000108653
|
tcaim
|
T-cell activation inhibitor, mitochondrial |
| chr20_-_18483106 | 0.07 |
ENSDART00000170864
|
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr4_-_2742712 | 0.07 |
|
|
|
| chr19_+_21783111 | 0.07 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr19_+_30210491 | 0.07 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr24_-_7558008 | 0.07 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr3_-_27470857 | 0.06 |
ENSDART00000077734
|
zgc:66160
|
zgc:66160 |
| chr3_+_27667194 | 0.06 |
ENSDART00000075100
|
carhsp1
|
calcium regulated heat stable protein 1 |
| chr10_+_33630200 | 0.05 |
ENSDART00000130093
|
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr15_-_5911297 | 0.04 |
|
|
|
| chr24_-_14446522 | 0.03 |
|
|
|
| chr12_-_20493683 | 0.03 |
|
|
|
| chr15_-_45446153 | 0.03 |
ENSDART00000100332
|
fgf12b
|
fibroblast growth factor 12b |
| chr10_+_28273581 | 0.02 |
ENSDART00000131003
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr16_-_13927947 | 0.02 |
ENSDART00000144856
|
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr6_+_3120600 | 0.02 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr4_-_2742799 | 0.01 |
|
|
|
| chr3_+_33608743 | 0.01 |
|
|
|
| chr2_+_50873893 | 0.01 |
|
|
|
| chr15_-_45304762 | 0.00 |
ENSDART00000177018
|
CABZ01068243.1
|
ENSDARG00000106883 |
| chr10_-_33677866 | 0.00 |
ENSDART00000142655
|
hunk
|
hormonally up-regulated Neu-associated kinase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 1.1 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |