Project
DANIO-CODE
Navigation
Downloads
Results for nr1d2a
Z-value: 0.36
Transcription factors associated with nr1d2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1d2a
|
ENSDARG00000003820 | nuclear receptor subfamily 1, group D, member 2a |
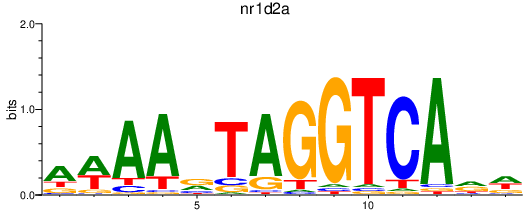
Activity profile of nr1d2a motif
Sorted Z-values of nr1d2a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1d2a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_19224936 | 0.39 |
ENSDART00000171103
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_48853800 | 0.37 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr14_-_40918015 | 0.32 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr2_-_56531858 | 0.31 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr7_+_65824459 | 0.31 |
ENSDART00000098259
ENSDART00000051823 ENSDART00000104523 |
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr16_-_24646282 | 0.30 |
ENSDART00000126274
|
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr21_+_30512356 | 0.30 |
ENSDART00000109719
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr19_+_19225148 | 0.29 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_39608334 | 0.29 |
ENSDART00000179059
|
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr20_-_49880744 | 0.27 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr19_+_19225228 | 0.26 |
ENSDART00000166172
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr21_-_12958161 | 0.25 |
ENSDART00000133517
|
MYORG
|
si:dkey-228b2.5 |
| chr7_-_29300448 | 0.25 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr2_+_58654570 | 0.25 |
ENSDART00000164102
ENSDART00000158777 |
cirbpa
|
cold inducible RNA binding protein a |
| chr22_-_38493856 | 0.24 |
ENSDART00000168676
|
soul4
|
heme-binding protein soul4 |
| chr23_+_44951868 | 0.24 |
|
|
|
| chr11_+_39768718 | 0.23 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr24_-_22611301 | 0.23 |
ENSDART00000169409
|
zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr22_-_817086 | 0.22 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr20_+_34417665 | 0.21 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr20_-_9992898 | 0.21 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr6_+_1895734 | 0.20 |
ENSDART00000109679
|
quo
|
quattro |
| chr20_-_4695108 | 0.19 |
ENSDART00000167224
ENSDART00000125620 ENSDART00000053858 |
papola
|
poly(A) polymerase alpha |
| chr8_+_14120313 | 0.18 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr21_-_24552672 | 0.18 |
|
|
|
| chr7_+_15619409 | 0.18 |
ENSDART00000144765
ENSDART00000145946 |
pax6b
|
paired box 6b |
| chr6_-_40314512 | 0.17 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr7_-_29300402 | 0.17 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr18_-_39491932 | 0.17 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr7_-_56472076 | 0.16 |
|
|
|
| chr18_-_35755677 | 0.16 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr3_+_39821041 | 0.16 |
|
|
|
| chr21_+_40082890 | 0.15 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr24_-_37407313 | 0.15 |
|
|
|
| chr4_-_22798521 | 0.15 |
ENSDART00000002851
ENSDART00000123801 ENSDART00000130409 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr15_-_35020312 | 0.14 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr3_+_32566693 | 0.13 |
ENSDART00000075244
|
fus
|
FUS RNA binding protein |
| chr2_+_58655334 | 0.13 |
|
|
|
| chr4_+_7668939 | 0.13 |
ENSDART00000149218
|
elk3
|
ELK3, ETS-domain protein |
| chr22_-_18520332 | 0.13 |
|
|
|
| chr9_-_49239691 | 0.13 |
ENSDART00000075627
|
cers6
|
ceramide synthase 6 |
| chr13_+_25356184 | 0.13 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr6_-_49160398 | 0.13 |
ENSDART00000143241
|
tspan2a
|
tetraspanin 2a |
| KN149955v1_+_4134 | 0.12 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr11_-_27832530 | 0.11 |
ENSDART00000172937
|
BX248318.1
|
ENSDARG00000104418 |
| chr19_+_5073991 | 0.11 |
ENSDART00000151050
|
ENSDARG00000011076
|
ENSDARG00000011076 |
| chr17_-_6580876 | 0.11 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr8_+_14120357 | 0.11 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr19_+_2659309 | 0.11 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr4_+_25296408 | 0.11 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr14_-_1081393 | 0.11 |
|
|
|
| chr18_-_15641660 | 0.10 |
ENSDART00000127614
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr8_+_43334181 | 0.10 |
ENSDART00000038566
|
fam101a
|
family with sequence similarity 101, member A |
| chr6_-_40771121 | 0.10 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr9_+_36051713 | 0.10 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr21_+_26660833 | 0.10 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| KN149955v1_+_4206 | 0.09 |
ENSDART00000167370
|
cdkn1d
|
cyclin-dependent kinase inhibitor 1D |
| chr3_-_15889742 | 0.09 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr1_+_19376127 | 0.09 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr5_-_22675495 | 0.08 |
ENSDART00000161883
|
si:dkey-114c15.5
|
si:dkey-114c15.5 |
| chr11_+_39768864 | 0.08 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr5_+_38143180 | 0.08 |
ENSDART00000137658
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr17_+_45892751 | 0.08 |
|
|
|
| chr7_-_14149889 | 0.08 |
ENSDART00000166519
|
kif7
|
kinesin family member 7 |
| chr10_-_8029671 | 0.08 |
ENSDART00000141445
|
ewsr1a
|
EWS RNA-binding protein 1a |
| chr10_-_7596917 | 0.08 |
ENSDART00000163689
|
wrn
|
Werner syndrome |
| chr9_-_37114484 | 0.08 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr25_-_17822363 | 0.08 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr21_-_2107554 | 0.08 |
ENSDART00000159378
|
AL627305.2
|
ENSDARG00000101832 |
| chr19_-_25842461 | 0.08 |
|
|
|
| chr2_+_33206349 | 0.07 |
ENSDART00000145588
|
rnf220a
|
ring finger protein 220a |
| chr17_-_29254289 | 0.07 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr1_+_48854316 | 0.07 |
|
|
|
| chr15_-_35020250 | 0.07 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr2_-_56896934 | 0.07 |
ENSDART00000164086
|
slc25a42
|
solute carrier family 25, member 42 |
| chr12_-_7201490 | 0.07 |
ENSDART00000152450
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr14_-_15776937 | 0.06 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr19_+_19224736 | 0.06 |
ENSDART00000158228
|
ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr11_-_21143897 | 0.06 |
ENSDART00000163008
|
RASSF5
|
Ras association domain family member 5 |
| chr4_+_18975027 | 0.06 |
ENSDART00000066973
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr5_-_68751287 | 0.06 |
ENSDART00000112692
|
ENSDARG00000077155
|
ENSDARG00000077155 |
| chr21_+_10698503 | 0.05 |
ENSDART00000144460
|
znf532
|
zinc finger protein 532 |
| chr19_-_10376154 | 0.05 |
ENSDART00000081485
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr16_-_22394481 | 0.05 |
|
|
|
| chr21_+_26569956 | 0.05 |
ENSDART00000100658
|
esrra
|
estrogen-related receptor alpha |
| chr5_+_38143502 | 0.05 |
ENSDART00000137658
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr3_+_57927416 | 0.05 |
ENSDART00000170144
|
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr25_-_27399421 | 0.05 |
ENSDART00000156906
|
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr12_+_23717554 | 0.04 |
ENSDART00000152997
|
svila
|
supervillin a |
| chr16_-_20084901 | 0.04 |
ENSDART00000103586
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr6_+_46968199 | 0.04 |
|
|
|
| chr9_-_10096542 | 0.04 |
ENSDART00000011922
|
spopla
|
speckle-type POZ protein-like a |
| chr18_-_35755825 | 0.04 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr3_+_52429876 | 0.04 |
ENSDART00000139037
|
pgls
|
6-phosphogluconolactonase |
| chr15_-_21733746 | 0.04 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr1_+_19282804 | 0.04 |
ENSDART00000170396
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr14_-_29518685 | 0.03 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr24_+_35173816 | 0.03 |
|
|
|
| chr3_+_34856339 | 0.03 |
ENSDART00000011319
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr21_+_10698081 | 0.03 |
ENSDART00000144460
|
znf532
|
zinc finger protein 532 |
| chr9_+_36051871 | 0.03 |
ENSDART00000144141
|
rcan1a
|
regulator of calcineurin 1a |
| chr6_-_45993454 | 0.03 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr16_+_30480096 | 0.03 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr17_-_42778156 | 0.03 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr23_-_35904414 | 0.03 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr8_+_2471513 | 0.03 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr12_+_19183576 | 0.03 |
ENSDART00000066391
|
csnk1e
|
casein kinase 1, epsilon |
| chr7_+_39957385 | 0.02 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr13_+_25356121 | 0.02 |
ENSDART00000057689
|
bag3
|
BCL2-associated athanogene 3 |
| chr18_-_15641870 | 0.02 |
ENSDART00000127614
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr12_-_7201358 | 0.01 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr13_+_18401965 | 0.01 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr21_-_10698972 | 0.01 |
|
|
|
| chr11_-_24300785 | 0.01 |
ENSDART00000171004
|
plekhg5a
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5a |
| chr17_-_15520620 | 0.01 |
ENSDART00000023320
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr20_+_36330995 | 0.00 |
ENSDART00000131867
|
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr21_-_1717752 | 0.00 |
|
|
|
| chr12_+_9504247 | 0.00 |
ENSDART00000127952
|
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr8_+_48106765 | 0.00 |
ENSDART00000086053
|
PRDM16
|
PR/SET domain 16 |
| chr20_-_49880706 | 0.00 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:1903428 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 0.2 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.0 | 0.1 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.0 | 0.2 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.3 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.3 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.0 | GO:0032585 | multivesicular body membrane(GO:0032585) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |