Project
DANIO-CODE
Navigation
Downloads
Results for nr1h4
Z-value: 0.67
Transcription factors associated with nr1h4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr1h4
|
ENSDARG00000057741 | nuclear receptor subfamily 1, group H, member 4 |
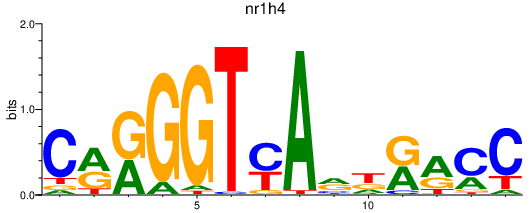
Activity profile of nr1h4 motif
Sorted Z-values of nr1h4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr1h4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_27697056 | 0.59 |
ENSDART00000132070
|
vamp5
|
vesicle-associated membrane protein 5 |
| chr1_-_11002715 | 0.56 |
ENSDART00000090689
|
brat1
|
BRCA1-associated ATM activator 1 |
| chr5_+_39885307 | 0.55 |
ENSDART00000051055
|
ndufs4
|
NADH dehydrogenase (ubiquinone) Fe-S protein 4, (NADH-coenzyme Q reductase) |
| chr23_+_22671069 | 0.54 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr23_+_28882409 | 0.52 |
ENSDART00000078171
|
pex14
|
peroxisomal biogenesis factor 14 |
| chr5_+_24569070 | 0.50 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr2_+_23004159 | 0.45 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr5_-_68799483 | 0.42 |
ENSDART00000145966
|
mapkapk5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr24_-_31394710 | 0.42 |
ENSDART00000169556
|
abcd3a
|
ATP-binding cassette, sub-family D (ALD), member 3a |
| chr25_-_16685648 | 0.42 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr18_+_46153484 | 0.40 |
ENSDART00000015034
|
blvrb
|
biliverdin reductase B |
| chr7_-_58856734 | 0.40 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr12_-_25110065 | 0.40 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr10_-_26240772 | 0.40 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr12_+_22449611 | 0.39 |
|
|
|
| chr14_+_10648299 | 0.39 |
ENSDART00000145776
|
zdhhc15b
|
zinc finger, DHHC-type containing 15b |
| chr7_-_58856862 | 0.38 |
ENSDART00000164104
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr22_-_11106940 | 0.37 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr21_+_11943637 | 0.36 |
ENSDART00000141306
|
zgc:162344
|
zgc:162344 |
| chr9_-_34459799 | 0.36 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr22_+_24532560 | 0.35 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr6_+_4068830 | 0.34 |
ENSDART00000130642
|
ENSDARG00000088225
|
ENSDARG00000088225 |
| chr15_+_29126271 | 0.33 |
ENSDART00000144546
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr11_+_24820942 | 0.32 |
|
|
|
| chr19_+_2754903 | 0.32 |
ENSDART00000160533
|
tomm7
|
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr5_-_68943600 | 0.31 |
ENSDART00000159851
|
mob1a
|
MOB kinase activator 1A |
| chr12_-_14104939 | 0.31 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr23_+_22671035 | 0.30 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr7_-_32708788 | 0.30 |
ENSDART00000113744
|
pkp3b
|
plakophilin 3b |
| chr23_-_42918086 | 0.29 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr21_-_2335024 | 0.28 |
ENSDART00000171645
|
si:ch73-299h12.9
|
si:ch73-299h12.9 |
| chr7_-_58856806 | 0.28 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr16_+_9635202 | 0.28 |
ENSDART00000148699
|
BX324207.1
|
ENSDARG00000095943 |
| chr23_+_4315077 | 0.28 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr5_-_68943821 | 0.28 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr17_-_17744885 | 0.27 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr5_+_1164747 | 0.26 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr2_-_6139833 | 0.26 |
ENSDART00000147509
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr12_-_26291790 | 0.26 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr24_+_26223767 | 0.26 |
|
|
|
| chr12_-_25110150 | 0.26 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr2_-_6139895 | 0.25 |
ENSDART00000147509
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr4_-_1809690 | 0.24 |
|
|
|
| chr14_+_22778880 | 0.24 |
ENSDART00000164526
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr2_-_37299150 | 0.24 |
ENSDART00000137598
|
nadkb
|
NAD kinase b |
| chr2_-_23003738 | 0.24 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr21_-_11943446 | 0.23 |
ENSDART00000142644
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr20_-_10500595 | 0.23 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr20_+_39782876 | 0.23 |
ENSDART00000050729
|
hddc2
|
HD domain containing 2 |
| chr12_-_18292591 | 0.23 |
ENSDART00000078724
|
atpaf2
|
ATP synthase mitochondrial F1 complex assembly factor 2 |
| chr21_+_11943609 | 0.23 |
ENSDART00000155426
ENSDART00000102463 |
zgc:162344
|
zgc:162344 |
| chr5_-_68664928 | 0.23 |
ENSDART00000073676
|
isca1
|
iron-sulfur cluster assembly 1 |
| chr15_+_46129807 | 0.22 |
ENSDART00000153936
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr2_+_10362661 | 0.22 |
ENSDART00000136018
|
nsun4
|
NOL1/NOP2/Sun domain family, member 4 |
| chr23_+_22671150 | 0.22 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr8_+_33010788 | 0.22 |
ENSDART00000172672
|
angptl2b
|
angiopoietin-like 2b |
| chr16_-_24646282 | 0.22 |
ENSDART00000126274
|
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr23_+_20505691 | 0.21 |
ENSDART00000140219
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr18_-_2989015 | 0.21 |
ENSDART00000159257
|
rsf1a
|
remodeling and spacing factor 1a |
| chr6_+_15018104 | 0.20 |
ENSDART00000063648
|
nck2b
|
NCK adaptor protein 2b |
| chr2_-_42384993 | 0.19 |
ENSDART00000141358
|
apom
|
apolipoprotein M |
| chr2_-_27996140 | 0.19 |
ENSDART00000097868
|
tgs1
|
trimethylguanosine synthase 1 |
| chr6_-_39633603 | 0.19 |
ENSDART00000104042
|
atf7b
|
activating transcription factor 7b |
| chr4_-_3340315 | 0.19 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr11_+_5845373 | 0.19 |
ENSDART00000111374
|
ndufs7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, (NADH-coenzyme Q reductase) |
| chr1_-_44239584 | 0.19 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr9_+_17415990 | 0.19 |
ENSDART00000112884
|
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr6_-_49512935 | 0.18 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr23_+_22313519 | 0.18 |
|
|
|
| chr5_+_40235387 | 0.18 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr6_-_19132923 | 0.18 |
|
|
|
| chr7_+_28881845 | 0.17 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr10_+_3728747 | 0.17 |
|
|
|
| chr21_-_2334989 | 0.16 |
ENSDART00000171645
|
si:ch73-299h12.9
|
si:ch73-299h12.9 |
| chr16_-_20506304 | 0.16 |
ENSDART00000134980
|
si:dkeyp-86h10.3
|
si:dkeyp-86h10.3 |
| chr2_+_27996280 | 0.16 |
ENSDART00000126762
|
tmem68
|
transmembrane protein 68 |
| chr12_-_18459868 | 0.16 |
|
|
|
| chr16_-_4940567 | 0.16 |
ENSDART00000149421
|
rpa2
|
replication protein A2 |
| chr7_+_38395803 | 0.16 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr2_+_38279454 | 0.16 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr21_-_32048768 | 0.16 |
ENSDART00000146079
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr19_-_27958005 | 0.15 |
ENSDART00000114301
|
si:ch211-152p11.4
|
si:ch211-152p11.4 |
| chr7_-_24567002 | 0.14 |
ENSDART00000139455
|
fam113
|
family with sequence similarity 113 |
| chr18_-_21199617 | 0.13 |
ENSDART00000060166
|
got2a
|
glutamic-oxaloacetic transaminase 2a, mitochondrial |
| chr10_-_26240909 | 0.13 |
ENSDART00000131394
|
fhdc3
|
FH2 domain containing 3 |
| chr18_+_31038010 | 0.13 |
ENSDART00000163471
|
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr11_-_18174 | 0.13 |
ENSDART00000167814
|
prr13
|
proline rich 13 |
| chr15_-_44054313 | 0.13 |
ENSDART00000110112
|
lamtor1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr15_+_30246345 | 0.13 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr5_+_65267043 | 0.13 |
ENSDART00000161578
|
mymk
|
myomaker, myoblast fusion factor |
| chr7_-_40359613 | 0.12 |
ENSDART00000173670
|
ube3c
|
ubiquitin protein ligase E3C |
| chr1_+_5948527 | 0.12 |
ENSDART00000138919
|
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr7_-_8635989 | 0.12 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr25_-_340711 | 0.11 |
ENSDART00000178866
|
CU929174.1
|
ENSDARG00000107546 |
| chr9_-_34459768 | 0.11 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr2_-_45838054 | 0.11 |
ENSDART00000125406
|
fam102ba
|
family with sequence similarity 102, member B, a |
| chr4_-_3340275 | 0.11 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr8_-_19311772 | 0.11 |
ENSDART00000138881
ENSDART00000010604 |
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr9_+_48309684 | 0.11 |
ENSDART00000177287
ENSDART00000113883 |
ENSDARG00000041339
|
ENSDARG00000041339 |
| chr22_-_38321005 | 0.11 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr18_+_17796754 | 0.10 |
|
|
|
| chr22_+_24291724 | 0.10 |
ENSDART00000168169
|
ccdc50
|
coiled-coil domain containing 50 |
| chr4_+_1680973 | 0.10 |
ENSDART00000149826
ENSDART00000166437 ENSDART00000031135 ENSDART00000172300 ENSDART00000067446 |
slc38a4
|
solute carrier family 38, member 4 |
| chr5_-_24568752 | 0.10 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr6_+_40954138 | 0.10 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr15_+_46093261 | 0.10 |
|
|
|
| chr3_+_48362748 | 0.10 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr6_-_586251 | 0.10 |
ENSDART00000148867
ENSDART00000149414 ENSDART00000149248 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr7_+_5837533 | 0.10 |
ENSDART00000110813
|
hist1h4l
|
histone 1, H4, like |
| chr13_-_7434672 | 0.10 |
ENSDART00000159453
|
h2afy2
|
H2A histone family, member Y2 |
| chr22_+_24291633 | 0.09 |
ENSDART00000165618
|
ccdc50
|
coiled-coil domain containing 50 |
| chr10_+_22043094 | 0.09 |
ENSDART00000035188
|
kcnip1b
|
Kv channel interacting protein 1 b |
| chr9_-_29832812 | 0.09 |
ENSDART00000101177
|
spryd7b
|
SPRY domain containing 7b |
| chr7_-_8635687 | 0.09 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr7_+_36267647 | 0.08 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr8_+_13757024 | 0.08 |
|
|
|
| chr22_-_10509388 | 0.08 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr5_+_35186037 | 0.08 |
ENSDART00000127383
ENSDART00000022043 |
stard8
|
StAR-related lipid transfer (START) domain containing 8 |
| chr12_+_34018417 | 0.08 |
ENSDART00000032821
|
cyth1b
|
cytohesin 1b |
| chr15_+_30246662 | 0.08 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr5_-_28090077 | 0.08 |
|
|
|
| chr7_+_29030304 | 0.07 |
ENSDART00000099327
|
rab8b
|
RAB8B, member RAS oncogene family |
| chr25_+_32062428 | 0.07 |
ENSDART00000152326
|
sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr8_+_7297420 | 0.07 |
ENSDART00000140874
|
seph
|
selenoprotein H |
| chr25_-_23949331 | 0.07 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr16_-_4940519 | 0.06 |
ENSDART00000149421
|
rpa2
|
replication protein A2 |
| chr16_+_5284778 | 0.06 |
ENSDART00000156685
ENSDART00000156765 |
soga3a
soga3a
|
SOGA family member 3a SOGA family member 3a |
| chr1_-_44002772 | 0.06 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr19_-_19282688 | 0.06 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr15_+_21231406 | 0.06 |
ENSDART00000038499
|
CR626907.1
|
ENSDARG00000077872 |
| chr8_+_45934564 | 0.06 |
ENSDART00000060955
|
nudt18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr20_-_49965261 | 0.05 |
|
|
|
| chr18_+_46153538 | 0.05 |
ENSDART00000114128
|
blvrb
|
biliverdin reductase B |
| chr1_+_127240 | 0.05 |
ENSDART00000122230
|
f7i
|
coagulation factor VIIi |
| chr4_-_18466436 | 0.05 |
ENSDART00000150221
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr8_+_13069019 | 0.05 |
ENSDART00000167926
|
itgb4
|
integrin, beta 4 |
| chr25_+_23494481 | 0.05 |
ENSDART00000089199
|
cpt1ab
|
carnitine palmitoyltransferase 1Ab (liver) |
| chr17_+_794430 | 0.04 |
ENSDART00000158830
|
cyp1c2
|
cytochrome P450, family 1, subfamily C, polypeptide 2 |
| chr13_-_25066650 | 0.04 |
|
|
|
| chr1_+_44239727 | 0.04 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr14_-_29542922 | 0.04 |
|
|
|
| chr15_+_24756002 | 0.04 |
ENSDART00000110618
|
LRRC75A
|
leucine rich repeat containing 75A |
| chr4_+_8531280 | 0.04 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr16_+_36465373 | 0.04 |
ENSDART00000158358
|
stk40
|
serine/threonine kinase 40 |
| chr23_+_20505072 | 0.04 |
ENSDART00000132920
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr6_-_31160132 | 0.04 |
ENSDART00000153592
|
CR847887.1
|
ENSDARG00000097226 |
| chr22_-_10509348 | 0.03 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr6_+_38629054 | 0.03 |
ENSDART00000086533
|
atp10a
|
ATPase, class V, type 10A |
| chr3_-_19750156 | 0.03 |
ENSDART00000134969
|
rnd2
|
Rho family GTPase 2 |
| chr3_+_31821397 | 0.03 |
ENSDART00000148861
|
kcnc3a
|
potassium voltage-gated channel, Shaw-related subfamily, member 3a |
| chr23_+_20505319 | 0.03 |
ENSDART00000140219
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr10_+_37033 | 0.03 |
ENSDART00000157786
|
riox2
|
ribosomal oxygenase 2 |
| chr3_-_32192499 | 0.03 |
|
|
|
| chr23_-_42918055 | 0.03 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr5_-_45986774 | 0.02 |
|
|
|
| chr4_+_8531350 | 0.02 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr22_+_24532479 | 0.02 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr10_+_16634200 | 0.02 |
ENSDART00000101142
|
chsy3
|
chondroitin sulfate synthase 3 |
| chr19_-_32487376 | 0.02 |
|
|
|
| chr14_-_33936261 | 0.02 |
ENSDART00000021437
|
gria1a
|
glutamate receptor, ionotropic, AMPA 1a |
| chr6_-_40314512 | 0.02 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr2_+_43757541 | 0.01 |
|
|
|
| chr12_+_16848206 | 0.01 |
|
|
|
| chr16_+_36465436 | 0.01 |
ENSDART00000158358
|
stk40
|
serine/threonine kinase 40 |
| chr15_-_30247084 | 0.00 |
ENSDART00000154069
|
BX322555.4
|
ENSDARG00000097908 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.3 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.2 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.4 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.1 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.0 | 0.1 | GO:0009155 | purine deoxyribonucleotide catabolic process(GO:0009155) |
| 0.0 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.6 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0014896 | regulation of muscle hypertrophy(GO:0014743) muscle hypertrophy(GO:0014896) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.5 | GO:0022900 | electron transport chain(GO:0022900) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 1.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.2 | GO:0048270 | methionine adenosyltransferase regulator activity(GO:0048270) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.3 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.0 | 0.4 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.7 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |