Project
DANIO-CODE
Navigation
Downloads
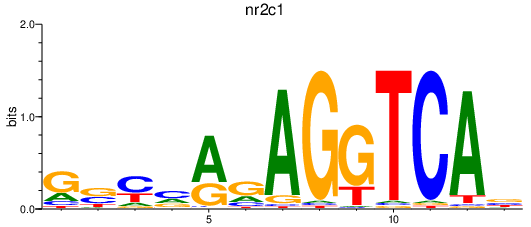
Results for nr2c1
Z-value: 0.28
Transcription factors associated with nr2c1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c1
|
ENSDARG00000045527 | nuclear receptor subfamily 2, group C, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c1 | dr10_dc_chr4_-_25869361_25869450 | -0.54 | 3.1e-02 | Click! |
Activity profile of nr2c1 motif
Sorted Z-values of nr2c1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_23463072 | 0.63 |
|
|
|
| chr10_-_21404605 | 0.50 |
ENSDART00000125167
|
avd
|
avidin |
| chr22_+_132285 | 0.49 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr4_-_17028492 | 0.42 |
|
|
|
| chr7_+_13742576 | 0.41 |
|
|
|
| chr11_+_37345493 | 0.36 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr10_-_180603 | 0.36 |
|
|
|
| chr7_+_13742622 | 0.36 |
|
|
|
| chr12_+_31623806 | 0.33 |
|
|
|
| chr10_+_15382647 | 0.33 |
ENSDART00000046274
|
trappc13
|
trafficking protein particle complex 13 |
| chr4_-_7867294 | 0.32 |
ENSDART00000167943
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr3_-_18655432 | 0.31 |
ENSDART00000034489
|
msrb1a
|
methionine sulfoxide reductase B1a |
| chr7_+_34025638 | 0.30 |
ENSDART00000052471
ENSDART00000173798 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr22_-_58352 | 0.29 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr11_+_24076334 | 0.29 |
ENSDART00000017599
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr11_+_24820942 | 0.29 |
|
|
|
| chr23_+_42793279 | 0.29 |
|
|
|
| chr15_+_29126271 | 0.29 |
ENSDART00000144546
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr3_-_58400454 | 0.28 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr25_+_32419008 | 0.27 |
|
|
|
| chr5_+_3683209 | 0.27 |
ENSDART00000132056
|
ggnbp2
|
gametogenetin binding protein 2 |
| chr3_-_32795702 | 0.27 |
ENSDART00000140117
|
aoc2
|
amine oxidase, copper containing 2 |
| chr6_-_31798839 | 0.26 |
|
|
|
| chr8_+_10825036 | 0.25 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr13_+_7243353 | 0.25 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr19_+_3115642 | 0.24 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_33362324 | 0.24 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr10_-_41018185 | 0.24 |
ENSDART00000134295
|
bmp1b
|
bone morphogenetic protein 1b |
| chr4_-_7867110 | 0.23 |
ENSDART00000162276
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr17_+_25828037 | 0.23 |
|
|
|
| chr2_-_41888389 | 0.22 |
|
|
|
| chr5_+_56658275 | 0.22 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr3_-_58400345 | 0.22 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr14_+_971438 | 0.22 |
ENSDART00000161487
|
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr4_-_951763 | 0.22 |
ENSDART00000093289
|
xrcc6bp1
|
XRCC6 binding protein 1 |
| chr24_-_12622124 | 0.21 |
|
|
|
| chr23_+_39454172 | 0.20 |
ENSDART00000149819
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr19_-_22761674 | 0.20 |
|
|
|
| chr3_+_27639256 | 0.20 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr11_-_37656066 | 0.20 |
|
|
|
| chr1_-_40536800 | 0.20 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr10_-_41018394 | 0.19 |
ENSDART00000059795
|
bmp1b
|
bone morphogenetic protein 1b |
| chr3_+_25904651 | 0.19 |
ENSDART00000078137
|
ankrd54
|
ankyrin repeat domain 54 |
| chr12_+_31668126 | 0.19 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr10_-_180681 | 0.19 |
|
|
|
| chr4_-_16461881 | 0.18 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr13_+_583640 | 0.18 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr21_+_2728070 | 0.18 |
|
|
|
| chr11_+_24076576 | 0.18 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_+_15382807 | 0.18 |
ENSDART00000168909
|
trappc13
|
trafficking protein particle complex 13 |
| chr19_+_3115514 | 0.18 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr5_-_45986774 | 0.18 |
|
|
|
| chr6_+_40525779 | 0.17 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr8_-_2471264 | 0.17 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr20_-_36768848 | 0.17 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr13_+_33545847 | 0.16 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr12_+_31668168 | 0.16 |
ENSDART00000105584
|
lrrc59
|
leucine rich repeat containing 59 |
| chr16_-_25765512 | 0.16 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr17_-_20110496 | 0.16 |
|
|
|
| chr19_-_3115104 | 0.16 |
ENSDART00000093146
|
bop1
|
block of proliferation 1 |
| chr8_+_10824974 | 0.16 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr3_-_33362384 | 0.16 |
ENSDART00000020342
|
sgsm3
|
small G protein signaling modulator 3 |
| chr13_+_7243325 | 0.15 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr16_-_25765396 | 0.15 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr24_+_14095601 | 0.15 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr23_+_39453929 | 0.15 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr5_+_26804676 | 0.15 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr8_+_21127033 | 0.15 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr16_-_25765322 | 0.15 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr13_+_6908853 | 0.15 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr25_-_27222696 | 0.14 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr5_-_41260390 | 0.14 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr15_+_40228295 | 0.14 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr9_+_33179226 | 0.14 |
|
|
|
| chr19_-_6503643 | 0.14 |
|
|
|
| chr3_+_40147509 | 0.14 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr2_+_32033176 | 0.13 |
ENSDART00000135040
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr23_+_37380022 | 0.13 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr5_+_56658431 | 0.13 |
ENSDART00000149646
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr12_+_19016508 | 0.13 |
ENSDART00000141346
ENSDART00000066397 |
phf5a
|
PHD finger protein 5A |
| chr16_+_36794641 | 0.12 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr3_+_19186724 | 0.12 |
ENSDART00000110548
|
kri1
|
KRI1 homolog |
| chr8_-_8659980 | 0.12 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr16_-_25765583 | 0.12 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr11_-_43125936 | 0.12 |
|
|
|
| chr11_+_24076371 | 0.12 |
ENSDART00000166045
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr21_-_43176006 | 0.11 |
ENSDART00000148630
|
hspa4a
|
heat shock protein 4a |
| chr24_+_17055519 | 0.11 |
ENSDART00000149954
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr3_-_36277628 | 0.11 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr23_+_22671035 | 0.11 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr20_+_28506477 | 0.11 |
ENSDART00000114929
|
zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr5_-_22090397 | 0.10 |
|
|
|
| chr3_+_19186494 | 0.10 |
ENSDART00000111528
|
kri1
|
KRI1 homolog |
| chr23_+_22671150 | 0.10 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr3_-_36277758 | 0.10 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr16_-_25765542 | 0.09 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr11_-_35757903 | 0.09 |
ENSDART00000167472
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr8_+_10824790 | 0.09 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr15_-_1880173 | 0.09 |
ENSDART00000154175
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr14_+_30073402 | 0.09 |
ENSDART00000136009
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr4_-_951970 | 0.09 |
ENSDART00000093289
|
xrcc6bp1
|
XRCC6 binding protein 1 |
| chr25_-_18910254 | 0.09 |
ENSDART00000019765
|
efcab6
|
EF-hand calcium binding domain 6 |
| chr1_-_51086075 | 0.09 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr9_-_41982745 | 0.09 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr16_-_25765623 | 0.08 |
ENSDART00000015302
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr7_-_65913414 | 0.08 |
ENSDART00000145843
|
pth1b
|
parathyroid hormone 1b |
| chr12_-_22387786 | 0.08 |
ENSDART00000109707
|
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr2_+_6155429 | 0.08 |
ENSDART00000145244
|
aldh9a1b
|
aldehyde dehydrogenase 9 family, member A1b |
| chr5_-_22090338 | 0.08 |
|
|
|
| chr5_-_30112312 | 0.08 |
|
|
|
| chr6_+_40954342 | 0.07 |
|
|
|
| chr25_+_18910528 | 0.07 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr11_+_3479741 | 0.07 |
ENSDART00000171621
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr17_+_16600773 | 0.07 |
|
|
|
| chr23_+_22671069 | 0.07 |
ENSDART00000054337
|
gpr157
|
G protein-coupled receptor 157 |
| chr17_+_28664644 | 0.07 |
|
|
|
| chr2_-_4276017 | 0.07 |
|
|
|
| chr10_-_20148585 | 0.07 |
ENSDART00000138120
ENSDART00000079922 |
pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr20_-_36768780 | 0.07 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr8_+_2471513 | 0.07 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr19_-_35117708 | 0.06 |
|
|
|
| chr17_-_49324003 | 0.06 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr3_+_53792981 | 0.06 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr9_+_17421457 | 0.05 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr1_+_54059634 | 0.05 |
ENSDART00000162075
|
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr6_+_40525610 | 0.05 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr12_+_26579286 | 0.05 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr15_+_28335293 | 0.05 |
ENSDART00000152536
|
myo1cb
|
myosin Ic, paralog b |
| chr16_+_8067533 | 0.05 |
ENSDART00000126041
|
ano10a
|
anoctamin 10a |
| chr7_+_41871444 | 0.04 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr2_-_4275952 | 0.04 |
|
|
|
| chr8_+_8660457 | 0.04 |
ENSDART00000137780
|
uxt
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr6_-_39905773 | 0.04 |
ENSDART00000148612
|
tsfm
|
Ts translation elongation factor, mitochondrial |
| chr3_+_25904908 | 0.04 |
ENSDART00000078137
|
ankrd54
|
ankyrin repeat domain 54 |
| chr9_+_38692733 | 0.04 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr20_+_38717757 | 0.04 |
ENSDART00000144521
|
si:ch211-243j20.2
|
si:ch211-243j20.2 |
| chr20_-_27038165 | 0.04 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr10_-_8238422 | 0.04 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr3_+_32539409 | 0.03 |
ENSDART00000136292
|
rnf25
|
ring finger protein 25 |
| chr6_-_51541332 | 0.03 |
ENSDART00000156336
ENSDART00000154512 |
si:dkey-6e2.2
|
si:dkey-6e2.2 |
| chr21_-_1956120 | 0.03 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr14_-_6818814 | 0.03 |
|
|
|
| chr7_+_31567140 | 0.03 |
ENSDART00000144679
|
mybpc3
|
myosin binding protein C, cardiac |
| chr18_-_16934961 | 0.03 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr17_-_22117248 | 0.02 |
ENSDART00000089689
|
mrps5
|
mitochondrial ribosomal protein S5 |
| chr11_+_6450876 | 0.02 |
ENSDART00000170180
|
CU062501.2
|
ENSDARG00000103967 |
| chr15_-_1880375 | 0.02 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr23_+_20184837 | 0.02 |
|
|
|
| chr1_-_43880612 | 0.02 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr13_-_37002066 | 0.02 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr15_+_40228075 | 0.02 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr24_+_30852751 | 0.02 |
|
|
|
| chr23_+_17946764 | 0.01 |
ENSDART00000117743
|
SNORD59
|
Small nucleolar RNA SNORD59 |
| chr15_+_36355348 | 0.01 |
|
|
|
| chr24_+_17055672 | 0.01 |
ENSDART00000149954
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr9_+_34623833 | 0.01 |
ENSDART00000175455
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr21_-_1956163 | 0.01 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr17_-_21348828 | 0.01 |
ENSDART00000058027
|
shtn1
|
shootin 1 |
| chr2_-_55970982 | 0.01 |
ENSDART00000154701
ENSDART00000154107 |
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr4_-_16461748 | 0.00 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr10_-_20545884 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098751 | bone cell development(GO:0098751) |
| 0.1 | 0.6 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.2 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.4 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.3 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.0 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.5 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.4 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.2 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.8 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.2 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0043878 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |