Project
DANIO-CODE
Navigation
Downloads
Results for nr2c2
Z-value: 0.88
Transcription factors associated with nr2c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2c2
|
ENSDARG00000042477 | nuclear receptor subfamily 2, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2c2 | dr10_dc_chr8_+_53241686_53241782 | 0.38 | 1.4e-01 | Click! |
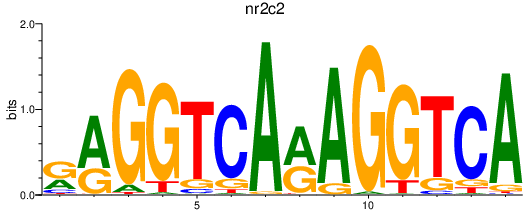
Activity profile of nr2c2 motif
Sorted Z-values of nr2c2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2c2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_12033088 | 1.57 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr5_-_36237656 | 1.51 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr13_-_39033893 | 1.47 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr5_-_21520111 | 1.32 |
|
|
|
| chr1_+_25157675 | 1.26 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr8_+_933677 | 1.24 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr3_+_39425125 | 1.18 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr3_-_55956030 | 1.18 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr2_+_42342148 | 1.18 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr9_+_33167554 | 1.07 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr9_+_21340251 | 1.02 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr13_+_48324283 | 1.01 |
ENSDART00000177067
|
CU929120.1
|
ENSDARG00000107321 |
| chr23_+_44817648 | 0.98 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr7_+_31567166 | 0.94 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr6_-_13654186 | 0.94 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr6_+_60060297 | 0.94 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr4_-_16461748 | 0.93 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr12_-_5085227 | 0.88 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr3_-_31672763 | 0.87 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr25_+_4490129 | 0.85 |
|
|
|
| chr22_-_17570306 | 0.84 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr21_-_18969970 | 0.79 |
ENSDART00000080269
|
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr10_-_44180588 | 0.79 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr3_-_24989269 | 0.76 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr19_-_19290260 | 0.76 |
|
|
|
| chr13_+_583640 | 0.76 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr5_-_1325831 | 0.75 |
|
|
|
| chr24_-_33817169 | 0.75 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr22_+_26543146 | 0.73 |
|
|
|
| chr17_+_15780370 | 0.72 |
ENSDART00000027667
ENSDART00000161637 |
rragd
|
ras-related GTP binding D |
| chr11_+_11217547 | 0.71 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr3_+_59560881 | 0.68 |
ENSDART00000084738
|
ppp1r27a
|
protein phosphatase 1, regulatory subunit 27a |
| chr15_+_8418722 | 0.68 |
ENSDART00000147086
|
tmem160
|
transmembrane protein 160 |
| chr9_-_41982635 | 0.60 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr12_+_27032862 | 0.59 |
|
|
|
| chr25_+_15551474 | 0.59 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr23_+_27985224 | 0.57 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr11_+_30482530 | 0.57 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr24_-_21027589 | 0.56 |
ENSDART00000154259
|
atp6v1ab
|
ATPase, H+ transporting, lysosomal, V1 subunit Ab |
| chr19_+_41274734 | 0.55 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr16_-_34304851 | 0.54 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr18_-_48498261 | 0.53 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr20_+_23643050 | 0.53 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr20_-_42805703 | 0.53 |
ENSDART00000045816
|
plg
|
plasminogen |
| chr20_+_2255204 | 0.53 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr2_-_43110857 | 0.53 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr20_+_15119519 | 0.52 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr16_-_1479139 | 0.52 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr20_-_22095097 | 0.51 |
ENSDART00000004984
|
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr19_+_11061965 | 0.51 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr13_+_28689749 | 0.50 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr19_-_32487376 | 0.48 |
|
|
|
| chr5_-_27394386 | 0.48 |
ENSDART00000171611
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr21_+_8106096 | 0.48 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr2_-_55737505 | 0.47 |
ENSDART00000059003
|
rx2
|
retinal homeobox gene 2 |
| chr5_-_47519273 | 0.47 |
|
|
|
| chr3_-_18261177 | 0.47 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr24_-_34449257 | 0.47 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr8_+_29751639 | 0.46 |
|
|
|
| chr13_+_6908853 | 0.45 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr20_+_15119754 | 0.44 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr17_+_11353143 | 0.43 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr23_-_624534 | 0.43 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr24_-_33817036 | 0.43 |
ENSDART00000079292
|
cavin4b
|
caveolae associated protein 4b |
| chr15_-_44957335 | 0.43 |
|
|
|
| chr6_-_40724581 | 0.42 |
ENSDART00000035101
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr19_+_32570656 | 0.42 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr9_+_33167471 | 0.42 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr16_+_5470419 | 0.42 |
|
|
|
| chr5_+_16669696 | 0.41 |
|
|
|
| chr4_-_17028492 | 0.41 |
|
|
|
| chr13_-_39033849 | 0.39 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr16_+_24697776 | 0.39 |
|
|
|
| chr14_+_721560 | 0.39 |
ENSDART00000165014
|
klb
|
klotho beta |
| chr24_-_23570982 | 0.38 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr9_+_11408481 | 0.38 |
ENSDART00000007308
|
wnt10a
|
wingless-type MMTV integration site family, member 10a |
| chr18_+_12452 | 0.38 |
|
|
|
| chr1_+_18909507 | 0.38 |
ENSDART00000088933
ENSDART00000141579 |
fbxo10
|
F-box protein 10 |
| chr17_+_443558 | 0.38 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr6_+_40953885 | 0.37 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr14_+_16508093 | 0.37 |
ENSDART00000161201
|
limch1b
|
LIM and calponin homology domains 1b |
| chr12_+_31345948 | 0.36 |
ENSDART00000031154
|
gucy2g
|
guanylate cyclase 2g |
| chr15_-_23532475 | 0.36 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr3_-_8608620 | 0.36 |
|
|
|
| chr5_-_70939686 | 0.36 |
|
|
|
| chr10_+_31358236 | 0.35 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr10_+_17490443 | 0.35 |
|
|
|
| chr3_-_18642744 | 0.35 |
ENSDART00000134208
|
hagh
|
hydroxyacylglutathione hydrolase |
| chr17_+_24018420 | 0.35 |
ENSDART00000140767
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr20_+_15264912 | 0.35 |
ENSDART00000063877
|
prdx6
|
peroxiredoxin 6 |
| chr1_+_43881022 | 0.35 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
| chr11_+_24720057 | 0.34 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr3_-_34050856 | 0.34 |
ENSDART00000151819
|
yipf2
|
Yip1 domain family, member 2 |
| chr20_+_27194162 | 0.34 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr13_-_1980818 | 0.34 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr21_+_302452 | 0.34 |
ENSDART00000151613
|
lhfpl2a
|
lipoma HMGIC fusion partner-like 2a |
| chr13_-_41420815 | 0.34 |
ENSDART00000163331
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr3_+_58096720 | 0.34 |
ENSDART00000010395
ENSDART00000159755 ENSDART00000171149 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein IIa |
| chr16_-_39181608 | 0.34 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr10_+_21693418 | 0.34 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr12_-_4648262 | 0.33 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr3_+_15027690 | 0.33 |
ENSDART00000065805
|
tspan10
|
tetraspanin 10 |
| KN149909v1_+_1319 | 0.33 |
|
|
|
| chr12_+_27370834 | 0.32 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr10_-_309863 | 0.32 |
ENSDART00000163287
|
ENSDARG00000102127
|
ENSDARG00000102127 |
| chr25_+_30701751 | 0.32 |
|
|
|
| chr24_-_21768269 | 0.32 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr9_-_1969197 | 0.32 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr9_-_48152388 | 0.31 |
|
|
|
| chr5_-_29916352 | 0.31 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr18_-_34573597 | 0.31 |
ENSDART00000021880
|
ssr3
|
signal sequence receptor, gamma |
| chr13_-_37001997 | 0.31 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr3_+_27639256 | 0.30 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr15_+_21231406 | 0.30 |
ENSDART00000038499
|
CR626907.1
|
ENSDARG00000077872 |
| chr25_+_21732255 | 0.30 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr20_+_25567046 | 0.30 |
ENSDART00000153071
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr25_+_28329427 | 0.30 |
ENSDART00000160619
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr25_+_18613868 | 0.30 |
ENSDART00000011149
|
fam185a
|
family with sequence similarity 185, member A |
| chr25_+_2909562 | 0.30 |
ENSDART00000149360
|
mpi
|
mannose phosphate isomerase |
| chr1_-_51658383 | 0.29 |
ENSDART00000132638
|
ENSDARG00000059948
|
ENSDARG00000059948 |
| chr10_-_17484971 | 0.29 |
|
|
|
| chr23_-_42918055 | 0.28 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr20_+_53716378 | 0.28 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr22_-_23641813 | 0.28 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr12_-_34612821 | 0.27 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr23_-_42918086 | 0.27 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr5_-_24568752 | 0.27 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr7_+_10346939 | 0.27 |
|
|
|
| chr23_+_42797015 | 0.27 |
|
|
|
| chr2_-_39052185 | 0.27 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr4_+_25296408 | 0.27 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr20_+_230164 | 0.26 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr15_-_29900 | 0.26 |
|
|
|
| chr10_-_29386085 | 0.26 |
ENSDART00000022605
|
tmem126a
|
transmembrane protein 126A |
| chr1_+_23093114 | 0.26 |
|
|
|
| chr23_+_8862155 | 0.26 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr5_-_18548689 | 0.26 |
ENSDART00000137802
|
fam214b
|
family with sequence similarity 214, member B |
| chr4_+_25296376 | 0.25 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr23_-_24307705 | 0.25 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr3_+_25868480 | 0.25 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr22_+_38276148 | 0.25 |
ENSDART00000104504
|
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr15_+_3296905 | 0.25 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr18_+_17622557 | 0.24 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr18_+_41323776 | 0.24 |
ENSDART00000048985
|
veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr12_-_31342432 | 0.24 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr24_-_23570895 | 0.24 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr7_+_36267647 | 0.24 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr22_-_16970979 | 0.24 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr22_+_18218979 | 0.24 |
|
|
|
| chr11_+_24720206 | 0.23 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr9_-_24309717 | 0.23 |
ENSDART00000140808
|
tyw5
|
tRNA-yW synthesizing protein 5 |
| chr8_-_386408 | 0.23 |
|
|
|
| chr9_+_32490721 | 0.23 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr8_+_7974156 | 0.23 |
|
|
|
| chr15_-_25431916 | 0.23 |
ENSDART00000047471
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr11_-_27531602 | 0.23 |
ENSDART00000023823
|
cass4
|
Cas scaffolding protein family member 4 |
| chr5_-_3103278 | 0.22 |
|
|
|
| chr6_+_28127444 | 0.22 |
|
|
|
| chr24_-_7558008 | 0.22 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr16_+_36794641 | 0.21 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr3_-_56474209 | 0.21 |
ENSDART00000156398
|
si:ch211-189a21.1
|
si:ch211-189a21.1 |
| chr18_-_7441136 | 0.21 |
ENSDART00000101250
|
si:dkey-30c15.13
|
si:dkey-30c15.13 |
| chr14_-_136402 | 0.21 |
|
|
|
| chr5_+_16669819 | 0.21 |
|
|
|
| chr23_+_37535830 | 0.20 |
|
|
|
| chr15_+_35105420 | 0.20 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr25_+_28450465 | 0.20 |
ENSDART00000075151
|
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr13_+_45843567 | 0.20 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr9_+_33311009 | 0.20 |
|
|
|
| chr2_-_11864995 | 0.20 |
ENSDART00000080925
|
impad1
|
inositol monophosphatase domain containing 1 |
| chr17_+_25463178 | 0.20 |
ENSDART00000139451
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr16_-_19762541 | 0.20 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr5_-_19987344 | 0.20 |
ENSDART00000144232
|
ficd
|
FIC domain containing |
| chr2_-_30005139 | 0.20 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr24_-_8365022 | 0.20 |
ENSDART00000149025
|
slc35b3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr7_+_31567140 | 0.19 |
ENSDART00000144679
|
mybpc3
|
myosin binding protein C, cardiac |
| chr14_-_44844086 | 0.19 |
ENSDART00000159988
|
fam193b
|
family with sequence similarity 193, member B |
| chr16_+_54906517 | 0.19 |
ENSDART00000126646
|
ENSDARG00000091079
|
ENSDARG00000091079 |
| chr21_+_30900987 | 0.18 |
ENSDART00000022562
|
rhogb
|
ras homolog family member Gb |
| chr1_-_35419335 | 0.18 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr16_-_24697750 | 0.18 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr12_+_26579396 | 0.18 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_-_7442720 | 0.18 |
ENSDART00000091105
|
mfap3l
|
microfibrillar-associated protein 3-like |
| chr16_+_39396358 | 0.18 |
|
|
|
| chr20_+_31375056 | 0.18 |
|
|
|
| chr20_-_10500595 | 0.18 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr23_+_44814525 | 0.17 |
|
|
|
| chr14_+_21999898 | 0.17 |
|
|
|
| chr13_-_37002066 | 0.17 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr17_+_16600773 | 0.17 |
|
|
|
| chr5_-_4483970 | 0.17 |
|
|
|
| chr10_+_31358307 | 0.17 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr7_-_55879197 | 0.17 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr22_-_16971180 | 0.17 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr12_+_27035744 | 0.16 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| KN150495v1_-_20497 | 0.16 |
|
|
|
| chr7_+_23735703 | 0.16 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr8_+_39768744 | 0.16 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr6_+_28127514 | 0.16 |
|
|
|
| chr5_-_18548376 | 0.16 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.3 | 1.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.9 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.2 | 1.8 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.5 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 1.8 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.3 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.1 | 0.3 | GO:0042671 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.2 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.3 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.8 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.5 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.3 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) chondrocyte morphogenesis(GO:0090171) |
| 0.1 | 0.6 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 0.2 | GO:0055091 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 1.2 | GO:0030388 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 1.6 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.5 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.8 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.8 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.4 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 2.5 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 1.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.3 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.4 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.2 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.2 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.5 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 1.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.0 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 0.9 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 1.8 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 1.6 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 1.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 2.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.9 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 0.9 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.8 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.1 | 0.5 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.0 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.0 | 0.5 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 0.5 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.7 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.7 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |