Project
DANIO-CODE
Navigation
Downloads
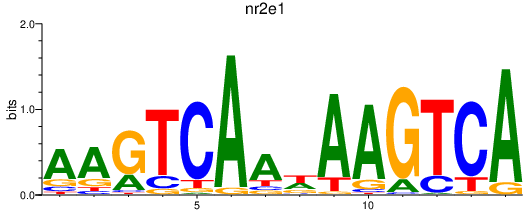
Results for nr2e1
Z-value: 0.37
Transcription factors associated with nr2e1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e1
|
ENSDARG00000017107 | nuclear receptor subfamily 2, group E, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2e1 | dr10_dc_chr20_-_32543497_32543565 | -0.17 | 5.2e-01 | Click! |
Activity profile of nr2e1 motif
Sorted Z-values of nr2e1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_31970637 | 0.91 |
ENSDART00000006124
|
cops2
|
COP9 signalosome subunit 2 |
| chr25_-_31979408 | 0.68 |
|
|
|
| chr12_-_28680035 | 0.63 |
ENSDART00000020667
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr21_+_6289363 | 0.43 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr7_+_58910115 | 0.29 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr17_+_33205732 | 0.29 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr1_+_57873295 | 0.29 |
ENSDART00000152744
|
CU550716.2
|
ENSDARG00000096528 |
| chr16_+_10538946 | 0.23 |
ENSDART00000173132
|
vars
|
valyl-tRNA synthetase |
| chr19_+_30210743 | 0.21 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr17_+_33205930 | 0.21 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr3_+_15605789 | 0.21 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr13_-_35766296 | 0.21 |
ENSDART00000162399
|
tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr17_+_33205888 | 0.20 |
ENSDART00000104476
|
snx9a
|
sorting nexin 9a |
| chr3_+_15605991 | 0.20 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr12_-_28680442 | 0.20 |
ENSDART00000178777
|
osbpl7
|
oxysterol binding protein-like 7 |
| chr22_-_22277386 | 0.20 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr23_+_19663746 | 0.18 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr2_-_48401640 | 0.18 |
ENSDART00000056277
|
gigfy2
|
GRB10 interacting GYF protein 2 |
| chr20_+_51193122 | 0.18 |
ENSDART00000028084
|
mpp5b
|
membrane protein, palmitoylated 5b (MAGUK p55 subfamily member 5) |
| chr17_+_38655239 | 0.17 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr11_-_18637646 | 0.17 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr19_+_30210491 | 0.16 |
ENSDART00000167803
|
marcksl1b
|
MARCKS-like 1b |
| chr6_-_52725402 | 0.16 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| KN150702v1_+_154982 | 0.16 |
ENSDART00000163214
|
CABZ01076670.1
|
ENSDARG00000104551 |
| chr23_+_28792316 | 0.16 |
ENSDART00000078156
|
srm
|
spermidine synthase |
| chr4_-_2937524 | 0.15 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr6_-_31377513 | 0.15 |
ENSDART00000145715
|
ak4
|
adenylate kinase 4 |
| chr10_-_10393735 | 0.14 |
|
|
|
| chr10_+_7634451 | 0.14 |
ENSDART00000162617
|
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr3_-_62156743 | 0.14 |
ENSDART00000101870
ENSDART00000175992 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr23_+_39961676 | 0.14 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| KN150711v1_+_9425 | 0.13 |
|
|
|
| chr12_-_33704092 | 0.13 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr25_-_13990944 | 0.13 |
ENSDART00000124140
|
zgc:101566
|
zgc:101566 |
| chr10_+_7634671 | 0.13 |
ENSDART00000171744
|
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr12_+_22552867 | 0.13 |
ENSDART00000152930
|
cdca9
|
cell division cycle associated 9 |
| chr13_-_6151100 | 0.13 |
ENSDART00000092105
|
agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| chr10_+_6925975 | 0.12 |
|
|
|
| chr1_+_9621488 | 0.12 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr3_+_15605866 | 0.12 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr1_+_9621612 | 0.12 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr19_+_30211052 | 0.11 |
ENSDART00000051804
|
marcksl1b
|
MARCKS-like 1b |
| chr13_-_37339532 | 0.11 |
ENSDART00000100324
|
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr2_-_55656088 | 0.11 |
ENSDART00000168579
|
CABZ01066725.1
|
ENSDARG00000100700 |
| chr25_+_35384119 | 0.11 |
ENSDART00000170551
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr25_-_12236333 | 0.11 |
ENSDART00000174863
ENSDART00000179042 |
BX323452.1
|
ENSDARG00000107774 |
| chr22_+_39067994 | 0.10 |
|
|
|
| chr18_+_13267740 | 0.10 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr5_-_29818204 | 0.10 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr13_-_37339582 | 0.10 |
ENSDART00000100324
|
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr3_+_42293213 | 0.10 |
ENSDART00000163595
|
ints1
|
integrator complex subunit 1 |
| chr18_+_13267658 | 0.10 |
ENSDART00000091560
|
tldc1
|
TBC/LysM-associated domain containing 1 |
| chr7_+_58910235 | 0.10 |
ENSDART00000172046
|
dok1b
|
docking protein 1b |
| chr7_+_58731472 | 0.10 |
ENSDART00000168888
|
fastkd3
|
FAST kinase domains 3 |
| chr13_+_24266204 | 0.10 |
ENSDART00000101200
ENSDART00000166005 |
zgc:153169
|
zgc:153169 |
| chr11_+_19271516 | 0.10 |
ENSDART00000162081
|
prickle2b
|
prickle homolog 2b |
| chr6_+_58628589 | 0.10 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr5_-_62982537 | 0.10 |
ENSDART00000111403
|
ciz1b
|
cdkn1a interacting zinc finger protein 1b |
| chr5_-_29818325 | 0.09 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr5_-_29818351 | 0.09 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr18_-_17796903 | 0.09 |
|
|
|
| chr13_+_24540979 | 0.09 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr13_-_37340209 | 0.09 |
|
|
|
| chr24_+_20514547 | 0.09 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr19_+_39689450 | 0.09 |
|
|
|
| chr7_+_68964813 | 0.09 |
ENSDART00000166258
|
marveld3
|
MARVEL domain containing 3 |
| chr1_-_51658383 | 0.09 |
ENSDART00000132638
|
ENSDARG00000059948
|
ENSDARG00000059948 |
| chr13_+_24266306 | 0.09 |
ENSDART00000101200
ENSDART00000166005 |
zgc:153169
|
zgc:153169 |
| chr24_+_20514498 | 0.08 |
ENSDART00000142848
|
sec22c
|
SEC22 homolog C, vesicle trafficking protein |
| chr6_-_40437353 | 0.08 |
ENSDART00000031153
|
ccdc174
|
coiled-coil domain containing 174 |
| chr19_-_11486168 | 0.08 |
ENSDART00000109889
ENSDART00000172065 |
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr7_+_23782982 | 0.08 |
|
|
|
| chr7_+_13130177 | 0.08 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr14_-_26078923 | 0.07 |
ENSDART00000022236
|
emx3
|
empty spiracles homeobox 3 |
| chr5_+_19506512 | 0.07 |
ENSDART00000047841
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr6_+_46404650 | 0.07 |
ENSDART00000131203
ENSDART00000103472 ENSDART00000132845 ENSDART00000138567 ENSDART00000168440 |
pbrm1l
|
polybromo 1, like |
| chr5_-_62982637 | 0.07 |
ENSDART00000111403
|
ciz1b
|
cdkn1a interacting zinc finger protein 1b |
| chr5_+_13370237 | 0.07 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr17_-_49931552 | 0.06 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr4_-_2937706 | 0.06 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr18_-_5689483 | 0.06 |
ENSDART00000129459
|
spata5l1
|
spermatogenesis associated 5-like 1 |
| chr3_+_30585864 | 0.06 |
|
|
|
| chr24_+_37839153 | 0.06 |
|
|
|
| chr21_+_21754830 | 0.06 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr1_+_13386937 | 0.06 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr16_+_36465436 | 0.05 |
ENSDART00000158358
|
stk40
|
serine/threonine kinase 40 |
| chr2_+_21947854 | 0.05 |
ENSDART00000112014
|
mplkip
|
M-phase specific PLK1 interacting protein |
| chr11_-_44895716 | 0.05 |
ENSDART00000170345
|
pfklb
|
phosphofructokinase, liver b |
| chr4_+_42865142 | 0.04 |
ENSDART00000150797
|
znf1073
|
zinc finger protein 1073 |
| chr7_+_13130111 | 0.04 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr4_-_39946152 | 0.04 |
|
|
|
| chr19_-_11486056 | 0.04 |
ENSDART00000109889
ENSDART00000172065 |
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr14_-_28228482 | 0.04 |
ENSDART00000166611
|
zgc:113364
|
zgc:113364 |
| chr5_-_34652798 | 0.04 |
ENSDART00000051278
|
tnpo1
|
transportin 1 |
| chr5_-_62187774 | 0.04 |
|
|
|
| chr16_+_36465373 | 0.04 |
ENSDART00000158358
|
stk40
|
serine/threonine kinase 40 |
| chr11_+_24076576 | 0.03 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr12_-_27497159 | 0.03 |
ENSDART00000066282
|
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr12_-_7669275 | 0.03 |
|
|
|
| chr20_+_27121126 | 0.03 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr14_+_31525014 | 0.03 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr7_-_65913414 | 0.03 |
ENSDART00000145843
|
pth1b
|
parathyroid hormone 1b |
| chr17_+_38655293 | 0.03 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr1_+_40763274 | 0.02 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr4_+_27139902 | 0.02 |
|
|
|
| chr3_+_15606121 | 0.02 |
ENSDART00000140160
|
BX914211.1
|
ENSDARG00000094116 |
| chr13_-_25153833 | 0.02 |
ENSDART00000039828
ENSDART00000110304 |
vcla
|
vinculin a |
| chr2_+_30266562 | 0.02 |
ENSDART00000135171
|
tmem70
|
transmembrane protein 70 |
| chr2_+_19339251 | 0.02 |
ENSDART00000172148
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr8_-_39850553 | 0.02 |
ENSDART00000131372
|
mlec
|
malectin |
| chr5_+_13370060 | 0.02 |
ENSDART00000160690
|
hk2
|
hexokinase 2 |
| chr9_+_21517603 | 0.02 |
|
|
|
| chr4_-_2937838 | 0.02 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr8_-_39850462 | 0.02 |
ENSDART00000131372
|
mlec
|
malectin |
| chr21_-_35291005 | 0.02 |
ENSDART00000134780
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr14_-_205130 | 0.02 |
ENSDART00000035581
ENSDART00000158725 ENSDART00000163549 |
OTOP1
|
otopetrin 1 |
| chr1_+_40762892 | 0.01 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr25_+_7391458 | 0.01 |
ENSDART00000042928
|
fuk
|
fucokinase |
| chr20_+_33391554 | 0.01 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr22_+_39067938 | 0.01 |
|
|
|
| chr24_-_25545773 | 0.01 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr16_+_54445145 | 0.01 |
ENSDART00000043403
|
ethe1
|
ethylmalonic encephalopathy 1 |
| chr5_+_60823823 | 0.01 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr20_+_38354928 | 0.01 |
ENSDART00000149160
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr9_-_23177842 | 0.01 |
|
|
|
| chr4_-_46037928 | 0.00 |
|
|
|
| chr5_+_42827955 | 0.00 |
ENSDART00000177895
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr4_+_59415357 | 0.00 |
ENSDART00000177097
|
BX255935.3
|
ENSDARG00000106194 |
| chr15_-_39756230 | 0.00 |
|
|
|
| chr16_+_27608837 | 0.00 |
ENSDART00000147611
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.2 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.0 | 0.2 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.1 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 0.0 | 0.2 | GO:0070252 | actin-mediated cell contraction(GO:0070252) |
| 0.0 | 0.1 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0003948 | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity(GO:0003948) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |