Project
DANIO-CODE
Navigation
Downloads
Results for nr2e3
Z-value: 1.83
Transcription factors associated with nr2e3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2e3
|
ENSDARG00000045904 | nuclear receptor subfamily 2, group E, member 3 |
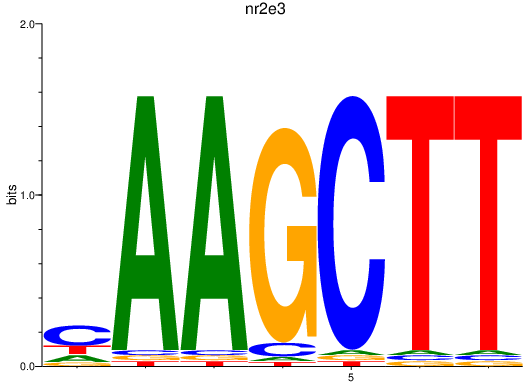
Activity profile of nr2e3 motif
Sorted Z-values of nr2e3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2e3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_47644421 | 6.81 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr19_+_41396014 | 5.46 |
ENSDART00000159444
ENSDART00000042990 ENSDART00000144544 |
col1a2
|
collagen, type I, alpha 2 |
| chr16_+_26900732 | 4.53 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr1_+_25662652 | 4.45 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr4_+_12033088 | 4.28 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr3_+_26896869 | 4.23 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr3_-_27516974 | 4.20 |
ENSDART00000151675
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr23_-_511984 | 4.00 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr10_+_4717800 | 3.93 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr18_+_43041488 | 3.91 |
|
|
|
| chr7_+_44373815 | 3.79 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr12_+_25509394 | 3.78 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr8_+_21163236 | 3.68 |
ENSDART00000091307
|
col2a1a
|
collagen, type II, alpha 1a |
| chr3_-_39313029 | 3.56 |
ENSDART00000156123
|
CU464118.1
|
ENSDARG00000097747 |
| chr18_+_40364675 | 3.51 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr20_+_4567653 | 3.42 |
|
|
|
| chr2_-_8219329 | 3.41 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr12_+_42281025 | 3.41 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr12_-_37560192 | 3.37 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr21_-_8103907 | 3.27 |
ENSDART00000171027
|
OLFML2A
|
olfactomedin like 2A |
| chr14_-_46651359 | 3.11 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr24_-_12794057 | 3.09 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr24_-_28814066 | 3.07 |
ENSDART00000042065
ENSDART00000170675 ENSDART00000158668 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr3_+_53518347 | 3.07 |
ENSDART00000170461
|
col5a3a
|
collagen, type V, alpha 3a |
| chr4_-_8610868 | 2.87 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr12_-_25796463 | 2.87 |
ENSDART00000152983
|
si:dkey-193p11.2
|
si:dkey-193p11.2 |
| chr4_-_73485204 | 2.81 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr9_+_25520011 | 2.75 |
ENSDART00000167110
|
itm2bb
|
integral membrane protein 2Bb |
| chr25_+_21732255 | 2.67 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr3_+_28450576 | 2.65 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr12_-_30662709 | 2.64 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr14_-_42633578 | 2.63 |
ENSDART00000162714
ENSDART00000161521 |
pcdh10b
|
protocadherin 10b |
| chr7_-_18302231 | 2.60 |
ENSDART00000108938
|
msantd1
|
Myb/SANT-like DNA-binding domain containing 1 |
| chr24_-_12794564 | 2.60 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_-_47842137 | 2.58 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr1_-_46052304 | 2.54 |
ENSDART00000142880
|
BX294379.1
|
ENSDARG00000092950 |
| chr24_-_34449257 | 2.53 |
ENSDART00000128690
|
agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr3_-_34671480 | 2.51 |
ENSDART00000167355
|
nsfa
|
N-ethylmaleimide-sensitive factor a |
| chr2_+_22324846 | 2.50 |
ENSDART00000044371
|
tox
|
thymocyte selection-associated high mobility group box |
| chr1_-_35419335 | 2.49 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr5_+_42406222 | 2.46 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr22_+_26543146 | 2.46 |
|
|
|
| chr19_+_156649 | 2.46 |
ENSDART00000167717
|
lrrc16a
|
leucine rich repeat containing 16A |
| chr12_+_31501522 | 2.46 |
ENSDART00000144422
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr7_+_21455138 | 2.45 |
ENSDART00000173992
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr1_-_22160662 | 2.44 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr5_+_38487183 | 2.44 |
ENSDART00000164065
|
anxa3a
|
annexin A3a |
| chr4_-_20435099 | 2.40 |
ENSDART00000055317
|
lrrc17
|
leucine rich repeat containing 17 |
| chr21_-_26459113 | 2.39 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| chr12_+_24833569 | 2.35 |
ENSDART00000014868
|
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr12_-_25973094 | 2.34 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr9_+_49054601 | 2.34 |
ENSDART00000047401
|
abcb11a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11a |
| chr24_+_19842162 | 2.32 |
ENSDART00000123031
|
CR381686.3
|
ENSDARG00000094073 |
| chr11_-_18542803 | 2.31 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr16_-_9978112 | 2.30 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr9_-_30437160 | 2.29 |
ENSDART00000147241
|
BX936337.1
|
ENSDARG00000092870 |
| chr5_-_33006537 | 2.23 |
ENSDART00000048350
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr8_-_15091823 | 2.22 |
ENSDART00000142358
|
bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr8_+_15989815 | 2.22 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr1_-_10822278 | 2.18 |
ENSDART00000091205
|
sdk1b
|
sidekick cell adhesion molecule 1b |
| chr19_+_20209561 | 2.18 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr14_-_46650724 | 2.15 |
|
|
|
| chr11_-_7310284 | 2.13 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr17_-_35934175 | 2.13 |
ENSDART00000110040
ENSDART00000137525 |
sox11a
AL929378.1
|
SRY (sex determining region Y)-box 11a ENSDARG00000092907 |
| chr24_-_26334717 | 2.12 |
ENSDART00000079984
ENSDART00000136871 |
rpl22l1
|
ribosomal protein L22-like 1 |
| chr23_-_28560598 | 2.11 |
ENSDART00000127072
|
tmem107l
|
transmembrane protein 107 like |
| chr2_+_36634309 | 2.10 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr1_+_46537108 | 2.10 |
|
|
|
| chr15_-_4537178 | 2.09 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr7_-_35438739 | 2.09 |
ENSDART00000043857
|
irx5a
|
iroquois homeobox 5a |
| chr3_-_32727588 | 2.08 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr24_+_24308055 | 2.06 |
|
|
|
| chr14_+_21563827 | 2.05 |
ENSDART00000032141
|
spon2a
|
spondin 2a, extracellular matrix protein |
| chr5_+_59845054 | 2.01 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr7_-_8128948 | 2.00 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr13_+_22528640 | 1.96 |
ENSDART00000078877
|
sncga
|
synuclein, gamma a |
| chr8_-_33372638 | 1.96 |
ENSDART00000076420
|
lmx1bb
|
LIM homeobox transcription factor 1, beta b |
| KN149932v1_+_27584 | 1.96 |
|
|
|
| chr17_-_14551031 | 1.96 |
ENSDART00000162452
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr17_+_134921 | 1.96 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr9_+_31471391 | 1.95 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr20_+_34679181 | 1.95 |
|
|
|
| chr3_+_36155364 | 1.95 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr17_+_27417635 | 1.93 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr7_+_63014869 | 1.92 |
ENSDART00000161436
|
pcdh7b
|
protocadherin 7b |
| chr7_-_46509291 | 1.92 |
ENSDART00000173891
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr22_+_18222691 | 1.88 |
ENSDART00000148357
|
BX664610.1
|
ENSDARG00000095557 |
| chr24_+_39266257 | 1.86 |
ENSDART00000053139
|
atp6v0cb
|
ATPase, H+ transporting, lysosomal, V0 subunit cb |
| chr20_-_9992898 | 1.86 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr18_+_40365208 | 1.86 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr5_+_37662896 | 1.86 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr5_-_62816208 | 1.85 |
ENSDART00000097325
|
c5
|
complement component 5 |
| chr20_+_35010140 | 1.84 |
ENSDART00000172001
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr25_-_12107407 | 1.83 |
ENSDART00000159800
ENSDART00000091727 |
ntrk3a
|
neurotrophic tyrosine kinase, receptor, type 3a |
| chr8_+_25748377 | 1.83 |
ENSDART00000062406
|
cacna1sb
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, b |
| chr19_-_22983970 | 1.82 |
ENSDART00000160153
|
pleca
|
plectin a |
| KN150170v1_+_10738 | 1.82 |
ENSDART00000159076
|
HUS1
|
HUS1 checkpoint clamp component |
| chr23_-_15807415 | 1.80 |
ENSDART00000178557
|
BX005011.3
|
ENSDARG00000108083 |
| chr1_-_18110990 | 1.80 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr19_-_22182031 | 1.78 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr7_-_14937467 | 1.77 |
ENSDART00000031049
|
ENSDARG00000023868
|
ENSDARG00000023868 |
| chr5_+_59844830 | 1.76 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr11_+_37639045 | 1.74 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr6_-_12079553 | 1.74 |
ENSDART00000157058
|
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr19_+_20202737 | 1.70 |
ENSDART00000164677
|
hoxa4a
|
homeobox A4a |
| chr15_+_14048904 | 1.70 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr7_+_19310071 | 1.70 |
ENSDART00000077545
|
slc7a7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr20_+_4567603 | 1.68 |
|
|
|
| chr9_-_22394674 | 1.67 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr3_-_34363698 | 1.67 |
|
|
|
| chr7_-_21978889 | 1.62 |
|
|
|
| chr4_+_22020246 | 1.62 |
ENSDART00000112035
ENSDART00000127664 |
myf5
|
myogenic factor 5 |
| chr16_+_5625301 | 1.62 |
|
|
|
| chr4_+_10722966 | 1.62 |
ENSDART00000102534
ENSDART00000067253 ENSDART00000136000 |
stab2
|
stabilin 2 |
| chr9_-_22377136 | 1.61 |
ENSDART00000110933
|
CR847505.1
|
ENSDARG00000076693 |
| chr14_+_23917724 | 1.61 |
ENSDART00000138082
ENSDART00000079215 |
stc2a
|
stanniocalcin 2a |
| chr1_-_11163480 | 1.60 |
ENSDART00000033361
|
ttyh3b
|
tweety family member 3b |
| chr21_+_22808694 | 1.60 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr17_+_48734060 | 1.56 |
ENSDART00000178697
|
daam2
|
dishevelled associated activator of morphogenesis 2 |
| chr1_-_53403731 | 1.56 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr3_-_28534446 | 1.56 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr24_-_12794672 | 1.55 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_-_35326559 | 1.55 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr16_+_53178713 | 1.54 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr21_-_20305406 | 1.54 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr5_+_68410884 | 1.53 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr19_-_22904687 | 1.53 |
ENSDART00000141503
|
pleca
|
plectin a |
| chr24_-_38899781 | 1.53 |
ENSDART00000112318
|
ntn2
|
netrin 2 |
| chr14_-_6822151 | 1.51 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr9_-_6683651 | 1.50 |
ENSDART00000061593
|
pou3f3a
|
POU class 3 homeobox 3a |
| chr20_+_13998066 | 1.48 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr4_-_73484546 | 1.48 |
|
|
|
| chr2_-_41669008 | 1.48 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr3_-_33295814 | 1.47 |
ENSDART00000146294
|
rpl23
|
ribosomal protein L23 |
| chr8_+_18552850 | 1.46 |
ENSDART00000177476
|
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr15_+_30276801 | 1.45 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr6_-_36574848 | 1.44 |
ENSDART00000135413
|
her6
|
hairy-related 6 |
| chr5_-_40451061 | 1.43 |
ENSDART00000083515
|
pdzd2
|
PDZ domain containing 2 |
| chr4_-_73485028 | 1.42 |
ENSDART00000122898
|
phf21b
|
PHD finger protein 21B |
| chr3_+_56008829 | 1.41 |
|
|
|
| chr17_+_18097490 | 1.41 |
ENSDART00000144894
|
bcl11ba
|
B-cell CLL/lymphoma 11Ba (zinc finger protein) |
| chr6_-_15514726 | 1.40 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr14_-_25301744 | 1.40 |
ENSDART00000131886
|
slc25a48
|
solute carrier family 25, member 48 |
| chr9_-_3700395 | 1.40 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr15_+_9351511 | 1.39 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr24_-_40968409 | 1.39 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr3_-_27517052 | 1.39 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr5_+_36180981 | 1.37 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr22_+_15480482 | 1.37 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr9_-_31714290 | 1.35 |
ENSDART00000127214
|
tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr3_+_15122138 | 1.35 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr1_-_11163282 | 1.35 |
ENSDART00000033361
|
ttyh3b
|
tweety family member 3b |
| chr1_+_25662910 | 1.35 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr23_+_24557956 | 1.34 |
|
|
|
| chr12_-_48758684 | 1.32 |
ENSDART00000130190
|
uros
|
uroporphyrinogen III synthase |
| chr3_+_49277125 | 1.32 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr3_+_23600462 | 1.30 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr17_+_48999061 | 1.30 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr16_-_21891429 | 1.29 |
ENSDART00000123597
|
ENSDARG00000089032
|
ENSDARG00000089032 |
| chr23_+_19743720 | 1.28 |
ENSDART00000031872
|
kctd6b
|
potassium channel tetramerization domain containing 6b |
| chr6_-_12953515 | 1.27 |
ENSDART00000150036
ENSDART00000149940 |
adam23a
|
ADAM metallopeptidase domain 23a |
| chr17_+_45961986 | 1.26 |
|
|
|
| chr5_+_51202141 | 1.26 |
ENSDART00000087467
|
cmya5
|
cardiomyopathy associated 5 |
| chr19_-_19029640 | 1.25 |
ENSDART00000169489
|
p3h4
|
prolyl 3-hydroxylase family member 4 (non-enzymatic) |
| chr13_+_30546398 | 1.23 |
|
|
|
| chr25_+_30535271 | 1.22 |
|
|
|
| chr17_-_27247075 | 1.22 |
|
|
|
| chr16_-_43441084 | 1.22 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr8_+_7297981 | 1.21 |
ENSDART00000170975
|
BX469901.2
|
ENSDARG00000103232 |
| chr15_+_15467483 | 1.19 |
ENSDART00000049831
|
dhrs11b
|
dehydrogenase/reductase (SDR family) member 11b |
| chr19_-_9603597 | 1.19 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr17_-_31041977 | 1.19 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr23_-_7283514 | 1.18 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr19_-_22984081 | 1.18 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr22_+_38276148 | 1.18 |
ENSDART00000104504
|
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr14_-_2245277 | 1.17 |
ENSDART00000169653
ENSDART00000160123 |
pcdh2ac
|
protocadherin 2 alpha c |
| chr16_+_5625552 | 1.17 |
|
|
|
| chr1_-_54319943 | 1.17 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr8_+_24725855 | 1.16 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr18_-_26483645 | 1.15 |
|
|
|
| chr9_+_33334773 | 1.15 |
ENSDART00000005879
|
atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr12_-_7902815 | 1.15 |
ENSDART00000088100
|
ank3b
|
ankyrin 3b |
| chr12_+_35912088 | 1.15 |
ENSDART00000167873
|
baiap2b
|
BAI1-associated protein 2b |
| chr7_-_35438887 | 1.14 |
ENSDART00000043857
|
irx5a
|
iroquois homeobox 5a |
| chr16_+_53111969 | 1.14 |
ENSDART00000011506
|
nkd2b
|
naked cuticle homolog 2b |
| chr25_-_325847 | 1.14 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr18_-_7423027 | 1.14 |
ENSDART00000147102
|
BX470218.1
|
ENSDARG00000094555 |
| chr15_+_18710214 | 1.14 |
ENSDART00000166555
|
cadm1b
|
cell adhesion molecule 1b |
| chr6_-_39278328 | 1.13 |
ENSDART00000148531
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr23_+_35996491 | 1.13 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr23_-_38228305 | 1.13 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr16_+_21891184 | 1.13 |
|
|
|
| chr5_+_18827478 | 1.12 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr6_-_40660116 | 1.12 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr3_+_23573114 | 1.12 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr9_+_34510213 | 1.10 |
ENSDART00000035522
ENSDART00000170615 ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr13_+_1484997 | 1.09 |
|
|
|
| chr3_+_36183975 | 1.09 |
ENSDART00000151305
|
slc16a6b
|
solute carrier family 16, member 6b |
| chr5_+_51201924 | 1.09 |
ENSDART00000087467
|
cmya5
|
cardiomyopathy associated 5 |
| chr10_-_22967786 | 1.08 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0071549 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 1.2 | 5.8 | GO:0019857 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.8 | 2.5 | GO:0048939 | Golgi vesicle docking(GO:0048211) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.8 | 3.3 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.6 | 3.7 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.6 | 1.7 | GO:0014005 | regulation of cellular amino acid metabolic process(GO:0006521) microglia development(GO:0014005) regulation of cellular amine metabolic process(GO:0033238) |
| 0.5 | 3.8 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.5 | 2.7 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.5 | 4.2 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.5 | 1.6 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.5 | 1.5 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.5 | 2.0 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.5 | 3.2 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.5 | 1.8 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.4 | 3.5 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.4 | 2.6 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.4 | 1.2 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.4 | 1.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.4 | 1.6 | GO:0060544 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.4 | 1.6 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.4 | 2.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.4 | 7.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 1.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 1.0 | GO:1990092 | calcium-dependent self proteolysis(GO:1990092) |
| 0.3 | 1.0 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 7.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.3 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 3.7 | GO:0021794 | thalamus development(GO:0021794) |
| 0.3 | 1.8 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.2 | 5.7 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.2 | 2.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 0.9 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.2 | 0.9 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 0.7 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 1.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.2 | 2.2 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 | 2.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 2.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 1.7 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 0.8 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.2 | 2.0 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.2 | 1.2 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 1.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 1.1 | GO:0044034 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 1.0 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.2 | 0.5 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.2 | 5.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 1.9 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 1.8 | GO:0090311 | regulation of protein deacetylation(GO:0090311) |
| 0.2 | 4.3 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.2 | 0.8 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 7.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 2.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.4 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.1 | 0.9 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.4 | GO:0034720 | negative regulation of ossification(GO:0030279) histone H3-K4 demethylation(GO:0034720) histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 1.1 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 0.7 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic G1 DNA damage checkpoint(GO:0031571) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 0.9 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 2.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 2.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.8 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.0 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 1.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.7 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 0.7 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 9.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.4 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.1 | 1.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 0.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 2.0 | GO:0021986 | habenula development(GO:0021986) |
| 0.1 | 4.2 | GO:0048596 | embryonic camera-type eye morphogenesis(GO:0048596) |
| 0.1 | 2.5 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 2.8 | GO:0010721 | negative regulation of cell development(GO:0010721) |
| 0.1 | 0.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.4 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.5 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.4 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.4 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 2.1 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 0.4 | GO:1900045 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 2.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.1 | 3.3 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.9 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 1.2 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.4 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.1 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.8 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 2.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 3.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 1.8 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.0 | 0.7 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.4 | GO:0051905 | establishment of pigment granule localization(GO:0051905) |
| 0.0 | 5.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.3 | GO:0031345 | negative regulation of cell projection organization(GO:0031345) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.3 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.2 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.0 | 1.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 1.0 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) positive regulation of vasculature development(GO:1904018) |
| 0.0 | 2.2 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0098773 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) skin epidermis development(GO:0098773) |
| 0.0 | 1.3 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 2.5 | GO:0002521 | leukocyte differentiation(GO:0002521) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.6 | GO:0042073 | intraciliary transport(GO:0042073) determination of ventral identity(GO:0048264) |
| 0.0 | 1.6 | GO:0006874 | cellular calcium ion homeostasis(GO:0006874) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 1.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.3 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.5 | GO:0006378 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.0 | 0.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.2 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 1.5 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 1.4 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.1 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.6 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.8 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.4 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 4.7 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 7.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.3 | 6.8 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.3 | 1.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 1.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 2.0 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 11.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.2 | 2.7 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 4.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 2.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.2 | 1.9 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 2.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.6 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 4.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 2.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 2.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 4.2 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 5.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 0.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.8 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 6.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 17.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.6 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 1.3 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 6.7 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.7 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.0 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 1.2 | 5.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.8 | 2.5 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.8 | 2.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 1.9 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.6 | 6.8 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.6 | 3.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.6 | 2.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.5 | 4.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.5 | 1.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.4 | 3.5 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 4.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.4 | 2.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.4 | 21.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 1.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.3 | 1.5 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.3 | 2.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 1.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.3 | 2.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 1.9 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.3 | 1.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.3 | 2.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 2.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 3.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 0.9 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.2 | 0.9 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.2 | 0.6 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 2.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 0.8 | GO:0046624 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.2 | 2.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 1.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 1.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.2 | 0.7 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.7 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 1.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.6 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 3.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 2.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 5.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 5.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 2.2 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 1.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.4 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 1.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.8 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 1.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.2 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.9 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 1.3 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 10.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.3 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 0.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 17.4 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 4.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 1.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 2.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 4.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 2.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 1.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.2 | 5.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.2 | 1.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 1.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 2.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.2 | 4.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 1.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.7 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 2.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.8 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.2 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |