Project
DANIO-CODE
Navigation
Downloads
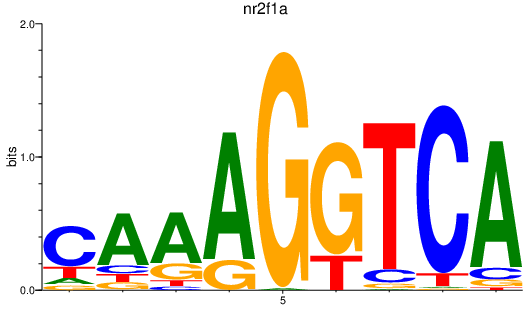
Results for nr2f1a
Z-value: 0.85
Transcription factors associated with nr2f1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1a
|
ENSDARG00000052695 | nuclear receptor subfamily 2, group F, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f1a | dr10_dc_chr5_+_49093134_49093226 | 0.86 | 1.6e-05 | Click! |
Activity profile of nr2f1a motif
Sorted Z-values of nr2f1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25157675 | 2.83 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr16_-_45943282 | 2.48 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr8_+_933677 | 2.35 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr20_+_31366832 | 2.21 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr22_-_17570306 | 2.11 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr5_+_40722565 | 2.11 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr12_-_5085227 | 1.99 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr16_-_45950530 | 1.95 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr17_+_30687093 | 1.86 |
ENSDART00000179434
|
apoba
|
apolipoprotein Ba |
| chr12_+_27032862 | 1.84 |
|
|
|
| chr22_+_15995914 | 1.69 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr3_+_39425125 | 1.67 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr5_+_8415025 | 1.67 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr25_+_4490129 | 1.62 |
|
|
|
| chr15_-_16948863 | 1.58 |
ENSDART00000062135
|
ENSDARG00000042379
|
ENSDARG00000042379 |
| chr9_-_39195468 | 1.55 |
ENSDART00000014207
|
myl1
|
myosin, light chain 1, alkali; skeletal, fast |
| chr2_-_42384993 | 1.50 |
ENSDART00000141358
|
apom
|
apolipoprotein M |
| chr12_+_13080209 | 1.50 |
ENSDART00000127870
|
cmn
|
calymmin |
| chr3_-_45779817 | 1.50 |
ENSDART00000164361
|
gcgra
|
glucagon receptor a |
| chr5_-_36237656 | 1.49 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr13_-_39033893 | 1.45 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr22_+_37934447 | 1.40 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr16_+_23999029 | 1.38 |
ENSDART00000129525
|
apoa4b.1
|
apolipoprotein A-IV b, tandem duplicate 1 |
| chr2_-_47578695 | 1.36 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr18_+_31038010 | 1.35 |
ENSDART00000163471
|
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr5_+_49093250 | 1.34 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr18_+_45893199 | 1.31 |
ENSDART00000158246
|
dvl3b
|
dishevelled segment polarity protein 3b |
| chr23_+_42488817 | 1.31 |
ENSDART00000171459
|
cyp2aa2
|
y cytochrome P450, family 2, subfamily AA, polypeptide 2 |
| chr13_-_37001997 | 1.29 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr6_+_48138737 | 1.27 |
|
|
|
| chr23_-_9924987 | 1.26 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr16_-_39181608 | 1.25 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr17_+_12254292 | 1.25 |
ENSDART00000160722
|
khk
|
ketohexokinase |
| chr20_-_36984259 | 1.24 |
ENSDART00000076313
|
txlnba
|
taxilin beta a |
| chr5_+_49093134 | 1.22 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr6_-_39315024 | 1.21 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr17_-_30685367 | 1.21 |
ENSDART00000114358
|
zgc:194392
|
zgc:194392 |
| chr24_-_14567403 | 1.19 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr9_-_34491458 | 1.19 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr22_+_30234718 | 1.19 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr21_-_20674965 | 1.18 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr3_-_31672763 | 1.17 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr9_+_42112576 | 1.16 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr11_+_6118409 | 1.15 |
ENSDART00000027666
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_+_22350043 | 1.14 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr3_-_18426055 | 1.12 |
ENSDART00000122968
|
aqp8b
|
aquaporin 8b |
| KN149909v1_+_1319 | 1.11 |
|
|
|
| chr16_+_21436660 | 1.11 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr11_+_21749658 | 1.09 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr17_+_1058137 | 1.08 |
|
|
|
| chr16_+_25491981 | 1.08 |
ENSDART00000086333
|
jarid2a
|
jumonji, AT rich interactive domain 2a |
| chr5_-_1325831 | 1.08 |
|
|
|
| chr11_+_26371444 | 1.06 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr19_+_156649 | 1.03 |
ENSDART00000167717
|
lrrc16a
|
leucine rich repeat containing 16A |
| chr22_+_24362712 | 1.03 |
ENSDART00000157861
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr23_-_15442032 | 1.03 |
ENSDART00000082060
|
ENSDARG00000078145
|
ENSDARG00000078145 |
| chr18_+_9413588 | 1.03 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr7_-_18894639 | 1.01 |
ENSDART00000142924
|
kirrel1a
|
kin of IRRE like a |
| chr25_+_5162045 | 1.01 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr9_+_9524547 | 1.00 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr11_+_11217547 | 1.00 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr14_+_28136958 | 0.99 |
|
|
|
| chr14_-_49115338 | 0.96 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr19_-_325337 | 0.96 |
ENSDART00000147635
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr2_+_32036450 | 0.96 |
ENSDART00000140776
|
CR391940.1
|
ENSDARG00000091946 |
| chr15_+_23849554 | 0.94 |
ENSDART00000138375
|
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr2_+_42342148 | 0.93 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr19_-_30816780 | 0.93 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr23_+_42381296 | 0.92 |
ENSDART00000158684
ENSDART00000159985 ENSDART00000172144 |
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr11_+_30482530 | 0.91 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr12_-_37921731 | 0.91 |
|
|
|
| chr23_+_36010592 | 0.91 |
ENSDART00000137507
|
hoxc3a
|
homeo box C3a |
| chr13_+_1377875 | 0.90 |
|
|
|
| chr20_+_2255204 | 0.89 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr23_+_24141576 | 0.89 |
|
|
|
| chr5_+_1223598 | 0.89 |
ENSDART00000159783
ENSDART00000026535 ENSDART00000168130 ENSDART00000147972 |
dnm1a
|
dynamin 1a |
| chr2_-_43110857 | 0.89 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr11_-_2107054 | 0.87 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr6_-_22227089 | 0.86 |
ENSDART00000168554
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr21_+_25199691 | 0.86 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr25_-_34638406 | 0.85 |
ENSDART00000123853
|
hist1h4l
|
histone 1, H4, like |
| chr20_+_23643050 | 0.85 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr9_+_9524688 | 0.84 |
ENSDART00000061525
ENSDART00000125174 |
nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr21_-_44013995 | 0.84 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr17_+_8874210 | 0.84 |
|
|
|
| chr12_-_3903961 | 0.82 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr10_-_32580373 | 0.81 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr1_-_50831155 | 0.81 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr2_+_37833161 | 0.81 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr4_-_21931540 | 0.80 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr14_+_35906366 | 0.80 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr4_-_20435099 | 0.79 |
ENSDART00000055317
|
lrrc17
|
leucine rich repeat containing 17 |
| chr12_-_31342432 | 0.79 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr23_+_44817648 | 0.79 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr14_-_48951428 | 0.78 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr9_-_1969197 | 0.77 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr15_+_28269289 | 0.77 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr16_-_32059848 | 0.77 |
ENSDART00000138701
|
gstk1
|
glutathione S-transferase kappa 1 |
| chr16_-_24753560 | 0.76 |
ENSDART00000063083
|
pon3.2
|
paraoxonase 3, tandem duplicate 2 |
| chr23_+_27985224 | 0.76 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr20_-_53162473 | 0.75 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr7_-_38288929 | 0.75 |
|
|
|
| KN150207v1_-_1208 | 0.75 |
ENSDART00000171112
|
CABZ01112575.1
|
ENSDARG00000099354 |
| chr24_+_16249188 | 0.75 |
ENSDART00000164516
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr8_+_7655655 | 0.74 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr14_-_16023198 | 0.74 |
|
|
|
| chr1_-_25600988 | 0.74 |
ENSDART00000160381
|
cxxc4
|
CXXC finger 4 |
| chr25_-_23949331 | 0.73 |
ENSDART00000047569
|
igf2b
|
insulin-like growth factor 2b |
| chr23_+_42431407 | 0.73 |
ENSDART00000169821
|
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr13_-_37144297 | 0.73 |
|
|
|
| chr13_-_31413855 | 0.73 |
ENSDART00000005670
|
dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr23_-_35657313 | 0.72 |
ENSDART00000043429
|
jph2
|
junctophilin 2 |
| chr5_+_26804344 | 0.72 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr19_+_4996328 | 0.71 |
ENSDART00000165082
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr22_-_17652938 | 0.71 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr13_-_219661 | 0.71 |
ENSDART00000133731
|
si:ch1073-291c23.2
|
si:ch1073-291c23.2 |
| chr2_-_59019569 | 0.71 |
|
|
|
| chr20_+_40247918 | 0.71 |
ENSDART00000121818
|
trdn
|
triadin |
| chr23_-_26798851 | 0.70 |
ENSDART00000170576
ENSDART00000166123 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr9_+_16437408 | 0.70 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr12_-_34726359 | 0.70 |
ENSDART00000153026
|
NDUFAF8
|
NADH:ubiquinone oxidoreductase complex assembly factor 8 |
| chr8_-_14146621 | 0.70 |
ENSDART00000063817
|
ndufb11
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11 |
| chr10_-_44180588 | 0.70 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr6_-_39767452 | 0.69 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_31567166 | 0.69 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr2_+_30932612 | 0.69 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr21_+_5483017 | 0.69 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr25_-_31881585 | 0.68 |
ENSDART00000123999
ENSDART00000158091 |
ENSDARG00000100731
|
ENSDARG00000100731 |
| KN150074v1_-_1031 | 0.68 |
|
|
|
| chr17_+_6119323 | 0.67 |
ENSDART00000131075
|
dusp23b
|
dual specificity phosphatase 23b |
| chr3_-_22082170 | 0.67 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr22_-_20101177 | 0.67 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr3_+_17913109 | 0.67 |
ENSDART00000112384
|
ankrd40
|
ankyrin repeat domain 40 |
| chr19_-_5416317 | 0.66 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr18_-_23889256 | 0.65 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr23_+_9285446 | 0.65 |
ENSDART00000033663
|
rps21
|
ribosomal protein S21 |
| chr16_+_26900732 | 0.65 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr9_-_21257082 | 0.64 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr16_+_30068125 | 0.64 |
ENSDART00000147083
|
CR847566.1
|
ENSDARG00000093232 |
| chr10_-_44441481 | 0.63 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr23_-_20494890 | 0.63 |
|
|
|
| chr5_-_29891016 | 0.63 |
ENSDART00000138464
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr9_+_21340251 | 0.63 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr18_+_27753985 | 0.63 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr18_+_7350133 | 0.63 |
ENSDART00000172404
|
cers3b
|
ceramide synthase 3b |
| chr11_+_30048913 | 0.63 |
|
|
|
| chr7_+_50491026 | 0.63 |
ENSDART00000165037
|
si:ch73-380l10.2
|
si:ch73-380l10.2 |
| chr8_+_25714744 | 0.62 |
ENSDART00000156340
|
si:ch211-167b20.8
|
si:ch211-167b20.8 |
| chr11_+_37345024 | 0.62 |
ENSDART00000077496
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr8_+_48488009 | 0.62 |
|
|
|
| chr3_-_24989269 | 0.62 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr20_+_18841440 | 0.62 |
ENSDART00000174532
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr12_+_23742199 | 0.62 |
|
|
|
| chr14_+_33073643 | 0.61 |
ENSDART00000052780
|
ndufa1
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 |
| KN150680v1_+_10854 | 0.61 |
|
|
|
| chr7_-_52574822 | 0.61 |
ENSDART00000172951
|
map1aa
|
microtubule-associated protein 1Aa |
| chr2_+_38057483 | 0.61 |
ENSDART00000159951
|
casq1a
|
calsequestrin 1a |
| chr4_-_1346961 | 0.61 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr23_-_24468191 | 0.61 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr2_+_29862841 | 0.61 |
ENSDART00000135918
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr15_-_25582891 | 0.61 |
|
|
|
| chr1_-_46015367 | 0.60 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr4_-_16461748 | 0.60 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr21_+_28441951 | 0.60 |
ENSDART00000077887
|
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr14_-_33596430 | 0.59 |
ENSDART00000112438
|
si:ch73-335m24.5
|
si:ch73-335m24.5 |
| chr3_-_6325652 | 0.59 |
ENSDART00000168508
|
FP236513.1
|
ENSDARG00000104099 |
| chr20_+_25567046 | 0.59 |
ENSDART00000153071
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr10_+_10393377 | 0.59 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr11_+_24720057 | 0.58 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr15_-_37973812 | 0.58 |
ENSDART00000122439
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr23_+_42370612 | 0.57 |
ENSDART00000161812
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr25_-_14948922 | 0.57 |
ENSDART00000161165
|
pax6a
|
paired box 6a |
| chr9_+_32490721 | 0.57 |
ENSDART00000078608
|
hspe1
|
heat shock 10 protein 1 |
| chr11_+_6118322 | 0.57 |
ENSDART00000165031
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr23_-_34970838 | 0.57 |
ENSDART00000087219
|
ENSDARG00000061272
|
ENSDARG00000061272 |
| chr25_-_22890657 | 0.57 |
|
|
|
| chr5_-_8568715 | 0.57 |
ENSDART00000099891
|
atp5ib
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit Eb |
| KN149795v1_-_24393 | 0.57 |
|
|
|
| chr11_-_30117563 | 0.57 |
ENSDART00000078387
|
pecr
|
peroxisomal trans-2-enoyl-CoA reductase |
| chr17_-_1111115 | 0.56 |
|
|
|
| chr20_+_230394 | 0.56 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr20_-_3970778 | 0.56 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr5_+_61335845 | 0.56 |
ENSDART00000166482
ENSDART00000050884 ENSDART00000163909 |
sepw1
|
selenoprotein W, 1 |
| chr20_+_4120302 | 0.56 |
ENSDART00000159837
|
cnksr3
|
cnksr family member 3 |
| chr4_-_25847471 | 0.56 |
ENSDART00000142491
|
ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr11_-_8158013 | 0.55 |
ENSDART00000024046
|
uox
|
urate oxidase |
| chr22_-_26971466 | 0.55 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr23_-_624534 | 0.55 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr21_+_8106096 | 0.55 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr16_+_21121428 | 0.55 |
|
|
|
| chr15_-_16271208 | 0.55 |
|
|
|
| chr10_+_26786051 | 0.55 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr11_+_21749918 | 0.55 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| KN150642v1_+_9419 | 0.55 |
ENSDART00000159069
|
atoh1b
|
atonal bHLH transcription factor 1b |
| chr9_-_22140954 | 0.55 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr23_+_32409339 | 0.55 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr18_-_15963983 | 0.54 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr17_-_31041977 | 0.54 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr8_-_23102555 | 0.54 |
|
|
|
| chr5_+_62304435 | 0.53 |
ENSDART00000123243
|
TIMM22
|
translocase of inner mitochondrial membrane 22 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0042671 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 1.7 | GO:0060843 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.3 | 2.6 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.3 | 0.9 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.3 | 3.6 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.3 | 3.9 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 0.5 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.3 | 0.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.3 | 0.8 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 0.8 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.2 | 1.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 1.1 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.6 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.2 | 0.8 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.2 | 1.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 1.5 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 0.9 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.5 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 0.7 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.2 | 0.5 | GO:0086010 | mechanosensory behavior(GO:0007638) membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.9 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 1.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.2 | 1.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 1.4 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 0.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.4 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 4.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 3.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.9 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.3 | GO:0097702 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.1 | 0.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.4 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.6 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 2.0 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.4 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 1.6 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.3 | GO:0030800 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.1 | 0.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.7 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.5 | GO:0006567 | glycine metabolic process(GO:0006544) threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:0015882 | L-ascorbic acid transport(GO:0015882) |
| 0.1 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.6 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.8 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.8 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.7 | GO:0042102 | positive regulation of mononuclear cell proliferation(GO:0032946) positive regulation of T cell proliferation(GO:0042102) positive regulation of lymphocyte proliferation(GO:0050671) positive regulation of leukocyte proliferation(GO:0070665) |
| 0.1 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.3 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 1.5 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.1 | 0.7 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.4 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.1 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.1 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.3 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.3 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.2 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.3 | GO:0035313 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.3 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0061036 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.4 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.8 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.2 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.2 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 1.8 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0060394 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 3.9 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.4 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.9 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.8 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 0.4 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.4 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.6 | GO:0071248 | cellular response to metal ion(GO:0071248) |
| 0.0 | 1.7 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.7 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.5 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.9 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.9 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.0 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.8 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 1.3 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 1.3 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 0.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.3 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.6 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.6 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.1 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.0 | 0.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.6 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 2.4 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.7 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.5 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.6 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.6 | 1.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 1.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 0.9 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.9 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 3.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 1.3 | GO:0044420 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 2.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 16.7 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.6 | 4.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.5 | 5.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.5 | 2.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.4 | 1.5 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.3 | 1.3 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.3 | 1.7 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 1.6 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.5 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.2 | 1.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.6 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.7 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.4 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 2.3 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 3.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.8 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.7 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.3 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.9 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.8 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.3 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.1 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 4.0 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.4 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.5 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.3 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 3.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 1.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.7 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 3.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 1.2 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.6 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |