Project
DANIO-CODE
Navigation
Downloads
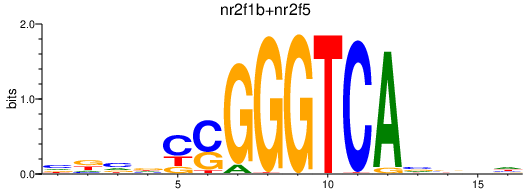
Results for nr2f1b+nr2f5
Z-value: 0.66
Transcription factors associated with nr2f1b+nr2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f1b
|
ENSDARG00000017168 | nuclear receptor subfamily 2, group F, member 1b |
|
nr2f5
|
ENSDARG00000033172 | nuclear receptor subfamily 2, group F, member 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f5 | dr10_dc_chr16_+_46327528_46327638 | 0.92 | 3.9e-07 | Click! |
| nr2f1b | dr10_dc_chr10_+_43950311_43950356 | 0.84 | 4.6e-05 | Click! |
Activity profile of nr2f1b+nr2f5 motif
Sorted Z-values of nr2f1b+nr2f5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f1b+nr2f5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_64175669 | 1.93 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr21_+_28408329 | 1.49 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr15_+_27431469 | 1.39 |
ENSDART00000122101
|
tbx2b
|
T-box 2b |
| chr15_-_27431216 | 1.25 |
ENSDART00000155933
|
AL954322.2
|
ENSDARG00000097072 |
| chr20_-_11179880 | 1.25 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr22_-_26971466 | 1.18 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr13_-_22877318 | 1.10 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr15_+_1238615 | 1.06 |
ENSDART00000167760
ENSDART00000163827 |
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr17_+_17935213 | 1.04 |
ENSDART00000104999
|
ccdc85ca
|
coiled-coil domain containing 85C, a |
| chr14_-_24094142 | 0.99 |
ENSDART00000126894
|
fam13b
|
family with sequence similarity 13, member B |
| KN149883v1_-_5164 | 0.98 |
ENSDART00000158506
ENSDART00000172360 |
NDUFA13
|
NADH:ubiquinone oxidoreductase subunit A13 |
| KN150642v1_+_9419 | 0.97 |
ENSDART00000159069
|
atoh1b
|
atonal bHLH transcription factor 1b |
| chr20_-_17006936 | 0.97 |
ENSDART00000012859
|
psma6b
|
proteasome subunit alpha 6b |
| chr6_-_39053400 | 0.96 |
ENSDART00000165839
|
tns2b
|
tensin 2b |
| chr2_+_33399405 | 0.96 |
ENSDART00000137207
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_53253623 | 0.96 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr10_-_21096635 | 0.93 |
ENSDART00000091004
|
pcdh1a
|
protocadherin 1a |
| chr1_-_55162455 | 0.92 |
ENSDART00000137909
|
samd1b
|
sterile alpha motif domain containing 1b |
| chr19_+_12315183 | 0.86 |
ENSDART00000149221
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr1_-_55108003 | 0.84 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr12_-_3742020 | 0.75 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr1_-_47080507 | 0.71 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr12_+_17744208 | 0.67 |
|
|
|
| chr21_+_25651712 | 0.66 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr13_+_24703802 | 0.66 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr3_-_12818954 | 0.65 |
ENSDART00000158747
ENSDART00000158815 |
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr8_-_41266057 | 0.64 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr5_+_64175494 | 0.59 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr16_+_21090083 | 0.56 |
ENSDART00000052662
|
hoxa13b
|
homeobox A13b |
| chr23_-_3760604 | 0.53 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr23_+_24010310 | 0.44 |
ENSDART00000171029
|
ptpn11b
|
protein tyrosine phosphatase, non-receptor type 11, b |
| chr4_+_1680973 | 0.44 |
ENSDART00000149826
ENSDART00000166437 ENSDART00000031135 ENSDART00000172300 ENSDART00000067446 |
slc38a4
|
solute carrier family 38, member 4 |
| chr11_+_43136945 | 0.44 |
|
|
|
| chr7_+_26980284 | 0.43 |
ENSDART00000173962
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr3_+_27615311 | 0.43 |
|
|
|
| chr2_+_58654570 | 0.41 |
ENSDART00000164102
ENSDART00000158777 |
cirbpa
|
cold inducible RNA binding protein a |
| chr1_-_47080468 | 0.37 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr4_-_70713033 | 0.35 |
ENSDART00000169130
|
ftr64
|
finTRIM family, member 64 |
| chr7_+_30803603 | 0.34 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr11_-_25086785 | 0.34 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr2_+_21947475 | 0.29 |
ENSDART00000112014
|
mplkip
|
M-phase specific PLK1 interacting protein |
| chr22_+_18291117 | 0.28 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr18_+_12452 | 0.27 |
|
|
|
| chr16_+_48729441 | 0.27 |
ENSDART00000154683
|
CU694532.1
|
ENSDARG00000097878 |
| chr14_-_5510157 | 0.26 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr20_+_46969915 | 0.25 |
ENSDART00000158124
|
ENSDARG00000103380
|
ENSDARG00000103380 |
| chr2_-_9809756 | 0.24 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr25_+_1511300 | 0.24 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr4_+_9010890 | 0.22 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr6_+_9441851 | 0.21 |
ENSDART00000064995
|
sumo1
|
small ubiquitin-like modifier 1 |
| chr5_-_24580478 | 0.18 |
ENSDART00000138209
|
dph7
|
diphthamide biosynthesis 7 |
| chr2_+_3062119 | 0.17 |
ENSDART00000110416
|
eif2b5
|
eukaryotic translation initiation factor 2B, subunit 5 epsilon |
| chr23_+_36494295 | 0.17 |
ENSDART00000042701
|
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr10_-_21096686 | 0.16 |
ENSDART00000139733
|
pcdh1a
|
protocadherin 1a |
| chr20_-_11179958 | 0.14 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr1_-_20131454 | 0.12 |
|
|
|
| chr12_+_26972001 | 0.11 |
|
|
|
| chr7_+_28881845 | 0.11 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr18_-_11707 | 0.11 |
ENSDART00000159781
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr12_+_26971596 | 0.11 |
|
|
|
| chr7_+_20829933 | 0.10 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr16_+_54024920 | 0.10 |
ENSDART00000155929
|
nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr15_-_35075545 | 0.10 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr21_+_5775383 | 0.08 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr1_-_39414732 | 0.08 |
ENSDART00000135647
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr20_+_53772078 | 0.08 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr15_+_2556736 | 0.04 |
ENSDART00000063329
|
cux1b
|
cut-like homeobox 1b |
| chr14_-_33468547 | 0.03 |
|
|
|
| chr19_+_27243907 | 0.03 |
ENSDART00000126006
|
FP085398.1
|
ENSDARG00000087611 |
| chr10_-_76359 | 0.00 |
ENSDART00000144722
|
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 1.0 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.2 | 0.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 1.5 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 1.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.2 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.9 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:1900006 | membrane tubulation(GO:0097320) positive regulation of dendrite development(GO:1900006) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0055004 | atrial cardiac myofibril assembly(GO:0055004) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 1.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 1.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |