Project
DANIO-CODE
Navigation
Downloads
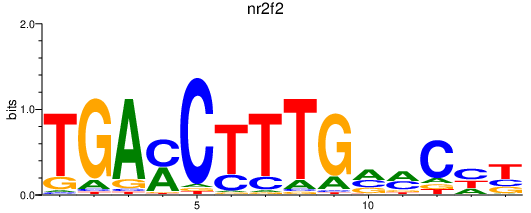
Results for nr2f2
Z-value: 0.66
Transcription factors associated with nr2f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f2
|
ENSDARG00000040926 | nuclear receptor subfamily 2, group F, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f2 | dr10_dc_chr18_-_23889169_23889233 | -0.73 | 1.4e-03 | Click! |
Activity profile of nr2f2 motif
Sorted Z-values of nr2f2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24964889 | 2.12 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr7_-_24249672 | 1.75 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr17_+_23463072 | 1.63 |
|
|
|
| chr22_-_38321005 | 1.36 |
ENSDART00000015117
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr4_-_16461881 | 1.35 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr22_+_132285 | 1.28 |
ENSDART00000059140
|
slc25a20
|
solute carrier family 25 (carnitine/acylcarnitine translocase), member 20 |
| chr19_-_3115104 | 1.26 |
ENSDART00000093146
|
bop1
|
block of proliferation 1 |
| chr17_-_2401636 | 1.24 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_-_73890499 | 1.20 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr11_+_29299382 | 1.18 |
|
|
|
| chr19_+_3115642 | 1.14 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr16_-_29452509 | 1.14 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr7_-_73890600 | 1.09 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr19_+_3115514 | 1.05 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr3_+_31530419 | 1.02 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr10_-_25808033 | 0.99 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr12_-_13692190 | 0.98 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr17_-_2396632 | 0.97 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr1_+_1564451 | 0.94 |
ENSDART00000137184
|
atp1a1a.2
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 2 |
| chr7_-_20358730 | 0.91 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr9_+_8402397 | 0.91 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr2_-_41888389 | 0.89 |
|
|
|
| chr9_+_33406198 | 0.88 |
ENSDART00000134029
ENSDART00000053061 |
si:ch211-125e6.12
si:ch211-125e6.13
|
si:ch211-125e6.12 si:ch211-125e6.13 |
| chr15_+_40228037 | 0.87 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr7_-_20358696 | 0.86 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr4_+_1770595 | 0.85 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr7_-_55879342 | 0.85 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr25_-_24105014 | 0.84 |
ENSDART00000048507
|
uevld
|
UEV and lactate/malate dehyrogenase domains |
| chr17_-_20159972 | 0.84 |
|
|
|
| chr9_+_41658126 | 0.82 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr2_+_23004159 | 0.80 |
ENSDART00000123442
|
zgc:161973
|
zgc:161973 |
| chr15_+_29126271 | 0.79 |
ENSDART00000144546
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr2_-_57020663 | 0.78 |
|
|
|
| chr7_-_24249630 | 0.77 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr23_-_33812361 | 0.77 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr18_-_20159345 | 0.76 |
|
|
|
| chr14_-_31149030 | 0.75 |
ENSDART00000018347
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr20_-_51843197 | 0.74 |
ENSDART00000129965
|
CABZ01079490.1
|
ENSDARG00000091595 |
| chr15_-_5913264 | 0.74 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr10_+_39272734 | 0.72 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr5_-_31796376 | 0.72 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr12_+_30257620 | 0.71 |
|
|
|
| chr15_-_30976483 | 0.70 |
ENSDART00000112511
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr7_-_24724911 | 0.69 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr2_-_57020490 | 0.69 |
|
|
|
| chr13_+_8364704 | 0.68 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr10_-_180603 | 0.67 |
|
|
|
| chr17_+_25828037 | 0.67 |
|
|
|
| chr25_-_36621626 | 0.67 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr10_+_15382647 | 0.67 |
ENSDART00000046274
|
trappc13
|
trafficking protein particle complex 13 |
| chr20_+_6545449 | 0.67 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr4_-_16461748 | 0.66 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr18_+_5066229 | 0.65 |
ENSDART00000157671
|
CABZ01080599.1
|
ENSDARG00000100626 |
| chr22_-_58352 | 0.65 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr18_+_44775449 | 0.65 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr24_-_2844245 | 0.63 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr1_-_40536800 | 0.63 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr14_+_971438 | 0.62 |
ENSDART00000161487
|
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr16_-_33047066 | 0.61 |
ENSDART00000147941
ENSDART00000075218 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr17_+_23908376 | 0.61 |
ENSDART00000021177
|
pex13
|
peroxisomal biogenesis factor 13 |
| chr10_-_25808063 | 0.61 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr8_-_22521204 | 0.61 |
ENSDART00000134542
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr5_+_3683209 | 0.60 |
ENSDART00000132056
|
ggnbp2
|
gametogenetin binding protein 2 |
| chr19_+_3115791 | 0.60 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr23_+_44814525 | 0.58 |
|
|
|
| chr19_+_3115685 | 0.58 |
ENSDART00000127473
ENSDART00000126549 ENSDART00000024593 ENSDART00000082353 ENSDART00000141324 |
hsf1
|
heat shock transcription factor 1 |
| chr3_-_18655432 | 0.58 |
ENSDART00000034489
|
msrb1a
|
methionine sulfoxide reductase B1a |
| chr15_+_40228295 | 0.55 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr6_-_19813235 | 0.55 |
ENSDART00000151152
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr11_+_26896056 | 0.55 |
ENSDART00000158411
ENSDART00000173374 ENSDART00000129736 |
hdac11
|
histone deacetylase 11 |
| chr7_-_20212038 | 0.54 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr3_-_23919508 | 0.54 |
ENSDART00000155896
|
BX936342.1
|
ENSDARG00000097621 |
| chr23_-_28881717 | 0.54 |
ENSDART00000078167
|
dffa
|
DNA fragmentation factor, alpha polypeptide |
| chr2_-_41722058 | 0.53 |
ENSDART00000022643
|
znf622
|
zinc finger protein 622 |
| chr4_-_17028492 | 0.53 |
|
|
|
| chr16_-_25318413 | 0.53 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr9_-_41352000 | 0.53 |
ENSDART00000059667
|
wdr75
|
WD repeat domain 75 |
| chr4_-_36838449 | 0.53 |
|
|
|
| chr10_-_39272682 | 0.52 |
ENSDART00000161758
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr4_+_1770198 | 0.51 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr9_+_29806055 | 0.50 |
ENSDART00000143493
|
phf11
|
PHD finger protein 11 |
| chr16_+_24045707 | 0.50 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr2_-_23003710 | 0.50 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr9_-_32873124 | 0.49 |
ENSDART00000041751
|
ercc1
|
excision repair cross-complementation group 1 |
| chr7_-_20358897 | 0.49 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr14_+_30073402 | 0.49 |
ENSDART00000136009
|
cnot7
|
CCR4-NOT transcription complex, subunit 7 |
| chr8_+_10825036 | 0.49 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr5_-_61654526 | 0.48 |
ENSDART00000158570
|
zgc:85789
|
zgc:85789 |
| chr2_+_44928188 | 0.48 |
ENSDART00000147292
|
ece2a
|
endothelin converting enzyme 2a |
| chr17_-_2419079 | 0.48 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr19_-_12276076 | 0.48 |
|
|
|
| chr14_+_21854812 | 0.48 |
ENSDART00000082697
ENSDART00000054982 |
gabra6a
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6a |
| chr4_+_1770500 | 0.47 |
ENSDART00000148486
|
scaf11
|
SR-related CTD-associated factor 11 |
| chr22_+_30553337 | 0.47 |
ENSDART00000139128
|
zgc:171679
|
zgc:171679 |
| chr15_+_40228382 | 0.46 |
|
|
|
| chr23_+_30803887 | 0.46 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr7_-_20212157 | 0.45 |
ENSDART00000078192
|
cnpy4
|
canopy4 |
| chr8_+_39768702 | 0.45 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr19_-_6503643 | 0.45 |
|
|
|
| chr25_+_17906515 | 0.45 |
|
|
|
| chr2_-_37116119 | 0.44 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr24_-_23570846 | 0.44 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr25_-_36621595 | 0.44 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr23_+_30803518 | 0.44 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr9_+_34571082 | 0.44 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr11_+_24820942 | 0.43 |
|
|
|
| chr6_-_8009143 | 0.43 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr15_+_40227972 | 0.43 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr7_-_20358841 | 0.42 |
ENSDART00000170422
|
si:dkey-19b23.8
|
si:dkey-19b23.8 |
| chr18_-_46185361 | 0.42 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr17_+_19711077 | 0.42 |
|
|
|
| chr15_-_1880173 | 0.42 |
ENSDART00000154175
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr5_+_62595346 | 0.42 |
ENSDART00000140065
|
si:ch73-37h15.2
|
si:ch73-37h15.2 |
| chr1_+_57938127 | 0.42 |
ENSDART00000158488
ENSDART00000172040 |
CABZ01084612.1
|
ENSDARG00000105295 |
| chr14_+_31149207 | 0.42 |
|
|
|
| chr17_-_49324003 | 0.42 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr14_+_268119 | 0.42 |
|
|
|
| chr5_+_62628554 | 0.42 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr18_-_16927749 | 0.42 |
ENSDART00000100105
ENSDART00000163400 |
swap70b
|
SWAP switching B-cell complex 70b subunit 70b |
| chr15_-_5912917 | 0.42 |
|
|
|
| chr3_+_40147509 | 0.41 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr5_-_31796734 | 0.41 |
ENSDART00000142095
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr23_-_28881880 | 0.41 |
ENSDART00000078167
|
dffa
|
DNA fragmentation factor, alpha polypeptide |
| chr13_-_30938624 | 0.41 |
ENSDART00000112653
|
wdfy4
|
WDFY family member 4 |
| chr25_-_36621405 | 0.41 |
ENSDART00000128108
|
urahb
|
urate (5-hydroxyiso-) hydrolase b |
| chr18_-_7488960 | 0.41 |
ENSDART00000052803
|
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr6_-_2037693 | 0.40 |
ENSDART00000159957
|
PXMP4
|
peroxisomal membrane protein 4 |
| chr10_-_180681 | 0.40 |
|
|
|
| chr17_+_19711104 | 0.40 |
|
|
|
| chr6_-_19812488 | 0.39 |
ENSDART00000151179
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr5_+_62611823 | 0.39 |
ENSDART00000177108
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr8_-_24194821 | 0.38 |
ENSDART00000177464
|
CT025875.1
|
ENSDARG00000106046 |
| chr7_-_55879197 | 0.38 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr5_-_34940744 | 0.37 |
ENSDART00000145779
|
si:dkey-204l11.1
|
si:dkey-204l11.1 |
| chr5_+_62682097 | 0.37 |
ENSDART00000137855
|
si:ch73-376l24.6
|
si:ch73-376l24.6 |
| chr1_+_38101290 | 0.37 |
ENSDART00000165405
|
spcs3
|
signal peptidase complex subunit 3 |
| chr16_-_25765512 | 0.37 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr16_-_25765396 | 0.36 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr8_-_8659980 | 0.36 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr9_+_18565169 | 0.36 |
|
|
|
| chr16_+_50355723 | 0.35 |
ENSDART00000166664
|
ENSDARG00000102916
|
ENSDARG00000102916 |
| chr9_+_30822402 | 0.35 |
|
|
|
| chr16_-_22378805 | 0.35 |
ENSDART00000124718
|
aqp10a
|
aquaporin 10a |
| chr24_-_30268889 | 0.35 |
ENSDART00000164187
|
snx7
|
sorting nexin 7 |
| chr5_-_41260390 | 0.35 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr16_-_25318334 | 0.34 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr6_-_33886299 | 0.34 |
ENSDART00000115409
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr16_-_25765322 | 0.34 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr7_+_19348229 | 0.33 |
ENSDART00000007310
ENSDART00000166355 |
zgc:171731
|
zgc:171731 |
| chr11_+_21452142 | 0.33 |
ENSDART00000170131
|
CR812468.1
|
ENSDARG00000100608 |
| chr2_+_54267946 | 0.33 |
ENSDART00000015535
|
ctnnbl1
|
catenin, beta like 1 |
| chr9_+_8919663 | 0.33 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr11_-_11591351 | 0.33 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr16_-_25318260 | 0.33 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr16_-_19762541 | 0.32 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr20_+_1100809 | 0.32 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr2_+_6155429 | 0.32 |
ENSDART00000145244
|
aldh9a1b
|
aldehyde dehydrogenase 9 family, member A1b |
| chr12_+_22459218 | 0.32 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr24_-_23570895 | 0.32 |
ENSDART00000084954
ENSDART00000004013 ENSDART00000129028 |
pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr6_+_55835894 | 0.32 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr2_+_54268010 | 0.31 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr20_+_6545194 | 0.31 |
ENSDART00000159829
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr17_+_45754797 | 0.31 |
ENSDART00000109525
|
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr25_-_19951695 | 0.31 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr9_+_19655550 | 0.31 |
ENSDART00000111973
|
pde9a
|
phosphodiesterase 9A |
| chr12_+_26579286 | 0.31 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_-_73890817 | 0.30 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr16_+_24046472 | 0.30 |
|
|
|
| chr16_+_36465436 | 0.30 |
ENSDART00000158358
|
stk40
|
serine/threonine kinase 40 |
| chr23_+_44814646 | 0.30 |
|
|
|
| chr17_-_20110496 | 0.29 |
|
|
|
| chr7_+_10188975 | 0.29 |
|
|
|
| chr16_+_55167489 | 0.29 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr8_+_10824974 | 0.29 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr12_-_49092133 | 0.29 |
|
|
|
| chr17_+_19711153 | 0.29 |
|
|
|
| chr11_-_36212977 | 0.29 |
ENSDART00000165203
|
usp48
|
ubiquitin specific peptidase 48 |
| chr1_+_1564283 | 0.28 |
ENSDART00000166968
|
atp1a1a.2
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 2 |
| chr22_+_20686657 | 0.28 |
|
|
|
| chr14_-_33468547 | 0.28 |
|
|
|
| chr10_+_24721086 | 0.28 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr16_-_25765542 | 0.28 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr17_+_21797726 | 0.28 |
ENSDART00000079011
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr17_-_2401568 | 0.28 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr8_+_21127033 | 0.28 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| KN150421v1_+_14181 | 0.28 |
|
|
|
| chr24_-_41403824 | 0.27 |
ENSDART00000063504
ENSDART00000169374 |
xylb
|
xylulokinase homolog (H. influenzae) |
| chr2_+_23600656 | 0.27 |
|
|
|
| chr10_+_20406983 | 0.27 |
ENSDART00000139022
|
golga7
|
golgin A7 |
| chr16_-_25765583 | 0.27 |
ENSDART00000132693
|
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr9_+_41658313 | 0.27 |
ENSDART00000100265
|
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr3_-_36277628 | 0.27 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr11_-_43125936 | 0.27 |
|
|
|
| chr22_+_20686400 | 0.27 |
|
|
|
| chr16_+_36465373 | 0.27 |
ENSDART00000158358
|
stk40
|
serine/threonine kinase 40 |
| chr5_-_68799310 | 0.26 |
ENSDART00000145966
|
mapkapk5
|
mitogen-activated protein kinase-activated protein kinase 5 |
| chr22_-_10744481 | 0.26 |
|
|
|
| chr13_-_42598533 | 0.26 |
ENSDART00000160472
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr6_+_40954342 | 0.26 |
|
|
|
| chr11_-_282662 | 0.26 |
ENSDART00000168461
|
poc1a
|
POC1 centriolar protein A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 1.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.2 | 1.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.2 | 3.0 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 1.0 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 0.5 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.4 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.1 | 3.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.1 | 0.3 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 1.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.3 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 1.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.2 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 1.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.5 | GO:0060078 | synaptic transmission, GABAergic(GO:0051932) regulation of postsynaptic membrane potential(GO:0060078) |
| 0.1 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 1.1 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.5 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.4 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) cGMP metabolic process(GO:0046068) |
| 0.0 | 0.1 | GO:0033146 | intracellular estrogen receptor signaling pathway(GO:0030520) regulation of intracellular estrogen receptor signaling pathway(GO:0033146) positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0071922 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.1 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 1.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.7 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 0.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.9 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.2 | 0.9 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.5 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.1 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0033971 | hydroxyisourate hydrolase activity(GO:0033971) |
| 0.3 | 1.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 3.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.9 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.2 | 0.6 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 0.6 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 1.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 1.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.6 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.3 | GO:0015168 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 1.8 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.1 | 0.3 | GO:0047105 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.1 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.3 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.2 | GO:0061609 | fructose-1-phosphate aldolase activity(GO:0061609) |
| 0.1 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 2.5 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.8 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.2 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 2.8 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 1.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.1 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.5 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |