Project
DANIO-CODE
Navigation
Downloads
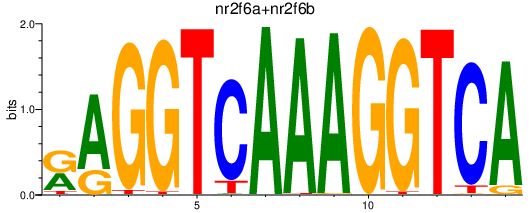
Results for nr2f6a+nr2f6b
Z-value: 0.85
Transcription factors associated with nr2f6a+nr2f6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr2f6b
|
ENSDARG00000003165 | nuclear receptor subfamily 2, group F, member 6b |
|
nr2f6a
|
ENSDARG00000003607 | nuclear receptor subfamily 2, group F, member 6a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr2f6b | dr10_dc_chr11_+_6118826_6118841 | 0.92 | 6.3e-07 | Click! |
| nr2f6a | dr10_dc_chr2_+_24718945_24718997 | 0.81 | 1.7e-04 | Click! |
Activity profile of nr2f6a+nr2f6b motif
Sorted Z-values of nr2f6a+nr2f6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr2f6a+nr2f6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_25157675 | 4.70 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr13_-_39033893 | 4.54 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr4_+_12033088 | 4.40 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr8_+_933677 | 4.12 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr22_+_26543146 | 3.30 |
|
|
|
| chr4_-_16461748 | 3.02 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr23_+_44817648 | 2.86 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr6_-_13654186 | 2.71 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr3_+_39425125 | 2.71 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr5_-_1325831 | 2.55 |
|
|
|
| chr15_-_35563367 | 2.27 |
|
|
|
| chr20_+_2255204 | 1.95 |
ENSDART00000137579
|
si:ch73-18b11.2
|
si:ch73-18b11.2 |
| chr5_+_49093250 | 1.87 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr11_+_11217547 | 1.80 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr9_-_41982635 | 1.76 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr5_+_49093134 | 1.69 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr8_+_29751639 | 1.56 |
|
|
|
| chr6_+_60060297 | 1.38 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr11_+_30482530 | 1.33 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr10_-_44180588 | 1.32 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr17_+_443558 | 1.29 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr5_-_47519273 | 1.16 |
|
|
|
| chr23_+_27985224 | 1.14 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr5_-_21520111 | 1.09 |
|
|
|
| chr1_+_23093114 | 1.07 |
|
|
|
| chr7_-_23775835 | 1.03 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr25_+_15551474 | 0.99 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr13_-_39033849 | 0.96 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr8_+_7974156 | 0.96 |
|
|
|
| chr16_+_55167489 | 0.95 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr16_-_50024848 | 0.94 |
ENSDART00000161782
|
etfb
|
electron-transfer-flavoprotein, beta polypeptide |
| chr13_-_37001997 | 0.90 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr9_-_48152388 | 0.88 |
|
|
|
| chr22_-_16971180 | 0.82 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr10_+_29966252 | 0.78 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr22_-_16970979 | 0.70 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr5_-_63031745 | 0.70 |
|
|
|
| chr22_-_16971035 | 0.64 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr12_-_31342432 | 0.64 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr6_+_28127444 | 0.59 |
|
|
|
| chr10_+_29965951 | 0.55 |
ENSDART00000116893
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr6_+_28127514 | 0.54 |
|
|
|
| chr6_+_42478185 | 0.53 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr13_+_45843567 | 0.52 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr15_+_6870469 | 0.52 |
|
|
|
| chr10_+_29966381 | 0.50 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr13_+_45843396 | 0.48 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr2_-_49060868 | 0.46 |
|
|
|
| chr12_+_26579396 | 0.43 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr24_-_21027589 | 0.35 |
ENSDART00000154259
|
atp6v1ab
|
ATPase, H+ transporting, lysosomal, V1 subunit Ab |
| chr13_-_37002066 | 0.34 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr12_+_26579286 | 0.32 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr8_+_39768744 | 0.29 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr10_+_29966533 | 0.27 |
ENSDART00000141549
|
hspa8
|
heat shock protein 8 |
| chr4_+_25868802 | 0.26 |
|
|
|
| chr9_+_33049445 | 0.24 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| KN150421v1_+_14181 | 0.24 |
|
|
|
| chr1_+_11485480 | 0.23 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr1_-_51086211 | 0.21 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr14_-_33468547 | 0.20 |
|
|
|
| chr11_+_30482504 | 0.18 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr25_+_21919595 | 0.17 |
ENSDART00000156517
|
ENSDARG00000062199
|
ENSDARG00000062199 |
| chr9_+_33049549 | 0.16 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr3_+_25868480 | 0.13 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr14_-_136402 | 0.12 |
|
|
|
| chr22_-_17570275 | 0.10 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr24_-_7558008 | 0.08 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr23_+_36241656 | 0.07 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr4_-_16461881 | 0.04 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr14_-_4117151 | 0.03 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.3 | 1.4 | GO:0019541 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.3 | 3.6 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.2 | 1.3 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.2 | 0.6 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 2.7 | GO:0001966 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 4.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.1 | 8.8 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 4.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.9 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 1.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.2 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 1.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 4.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 5.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 8.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.5 | 4.4 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.3 | 2.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 4.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 3.6 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.9 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 2.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |