Project
DANIO-CODE
Navigation
Downloads
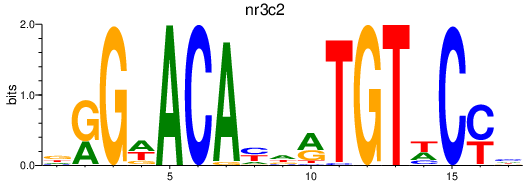
Results for nr3c2
Z-value: 0.44
Transcription factors associated with nr3c2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr3c2
|
ENSDARG00000102082 | nuclear receptor subfamily 3, group C, member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr3c2 | dr10_dc_chr1_-_36356001_36356009 | -0.74 | 9.8e-04 | Click! |
Activity profile of nr3c2 motif
Sorted Z-values of nr3c2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr3c2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_50452131 | 1.36 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr6_-_46740359 | 1.22 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr20_-_14885629 | 1.19 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr23_+_27777325 | 1.08 |
ENSDART00000134008
|
lmbr1l
|
limb development membrane protein 1-like |
| chr20_-_14885599 | 1.03 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr18_-_20619155 | 0.95 |
ENSDART00000151864
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr6_+_41098443 | 0.93 |
ENSDART00000143741
|
fkbp5
|
FK506 binding protein 5 |
| chr5_-_9036049 | 0.89 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
| chr6_+_277714 | 0.86 |
|
|
|
| chr6_+_277946 | 0.78 |
|
|
|
| chr18_-_20618924 | 0.73 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr15_+_40340914 | 0.69 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr14_-_47125993 | 0.68 |
ENSDART00000124925
|
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr22_-_35003371 | 0.64 |
ENSDART00000133537
|
arhgap19
|
Rho GTPase activating protein 19 |
| chr5_-_9035789 | 0.63 |
ENSDART00000124384
|
gak
|
cyclin G associated kinase |
| chr23_+_27777111 | 0.62 |
ENSDART00000134008
|
lmbr1l
|
limb development membrane protein 1-like |
| chr8_+_39734109 | 0.57 |
ENSDART00000017153
|
hps4
|
Hermansky-Pudlak syndrome 4 |
| chr24_-_23639458 | 0.54 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr24_-_23639325 | 0.49 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr6_+_50452229 | 0.45 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr19_+_33551956 | 0.42 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr11_+_5883229 | 0.40 |
ENSDART00000129663
|
dazap1
|
DAZ associated protein 1 |
| chr7_-_51474531 | 0.39 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr13_-_33348231 | 0.39 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr24_-_23639592 | 0.38 |
ENSDART00000090368
|
sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr5_-_54169271 | 0.36 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr3_-_25144722 | 0.35 |
ENSDART00000055445
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr11_+_5883270 | 0.33 |
ENSDART00000130768
|
dazap1
|
DAZ associated protein 1 |
| chr9_+_24084807 | 0.31 |
ENSDART00000131953
|
cops8
|
COP9 signalosome subunit 8 |
| chr5_+_18837448 | 0.31 |
|
|
|
| chr5_+_26205999 | 0.30 |
ENSDART00000111019
|
rabl6b
|
RAB, member RAS oncogene family-like 6b |
| chr8_+_21127033 | 0.30 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr18_-_44617805 | 0.30 |
ENSDART00000173095
ENSDART00000136420 |
spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr8_-_25889113 | 0.23 |
|
|
|
| chr10_+_29963102 | 0.20 |
ENSDART00000168898
|
hspa8
|
heat shock protein 8 |
| chr20_+_24047842 | 0.19 |
ENSDART00000128616
ENSDART00000144195 |
casp8ap2
|
caspase 8 associated protein 2 |
| chr18_-_19501985 | 0.15 |
ENSDART00000090413
|
snapc5
|
small nuclear RNA activating complex, polypeptide 5 |
| chr6_-_27067072 | 0.14 |
ENSDART00000149363
|
stk25a
|
serine/threonine kinase 25a |
| chr25_-_20993910 | 0.11 |
|
|
|
| chr7_-_16345978 | 0.10 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr5_+_29918213 | 0.10 |
ENSDART00000161836
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr10_-_1690504 | 0.07 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr24_-_36248243 | 0.07 |
ENSDART00000065336
|
tmem14cb
|
transmembrane protein 14Cb |
| chr5_-_9035835 | 0.04 |
ENSDART00000160079
|
gak
|
cyclin G associated kinase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.3 | 2.2 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 1.7 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 1.7 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.7 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.9 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |