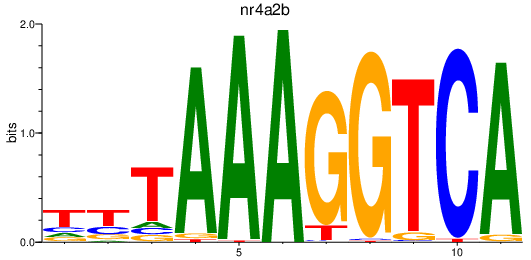
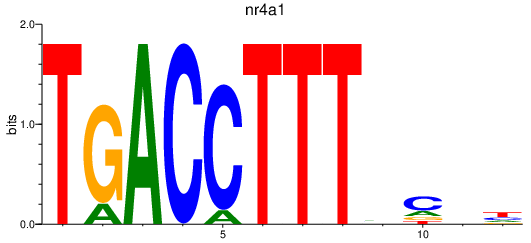
Project
DANIO-CODE
Navigation
Downloads
Results for nr4a2b_nr4a1
Z-value: 0.70
Transcription factors associated with nr4a2b_nr4a1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr4a2b
|
ENSDARG00000044532 | nuclear receptor subfamily 4, group A, member 2b |
|
nr4a1
|
ENSDARG00000000796 | nuclear receptor subfamily 4, group A, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr4a1 | dr10_dc_chr23_-_32231333_32231345 | 0.64 | 7.5e-03 | Click! |
Activity profile of nr4a2b_nr4a1 motif
Sorted Z-values of nr4a2b_nr4a1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr4a2b_nr4a1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_39315024 | 3.08 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr22_+_30234718 | 2.83 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr5_+_1223598 | 2.42 |
ENSDART00000159783
ENSDART00000026535 ENSDART00000168130 ENSDART00000147972 |
dnm1a
|
dynamin 1a |
| chr18_+_9413588 | 2.35 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr25_+_4490129 | 1.92 |
|
|
|
| chr3_+_39425125 | 1.90 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr7_-_18894639 | 1.66 |
ENSDART00000142924
|
kirrel1a
|
kin of IRRE like a |
| chr5_-_1325831 | 1.64 |
|
|
|
| chr13_+_1377875 | 1.52 |
|
|
|
| chr14_-_16815986 | 1.49 |
ENSDART00000060479
|
smtnl1
|
smoothelin-like 1 |
| chr20_+_40247918 | 1.43 |
ENSDART00000121818
|
trdn
|
triadin |
| chr16_+_24057260 | 1.42 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr4_-_16461748 | 1.38 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr14_-_16816075 | 1.37 |
ENSDART00000060479
|
smtnl1
|
smoothelin-like 1 |
| chr25_+_18866796 | 1.36 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr3_+_23580269 | 1.26 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr5_+_40722565 | 1.22 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr22_-_17570306 | 1.20 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr11_-_29908609 | 1.20 |
ENSDART00000156196
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr9_-_21257082 | 1.11 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr6_+_48138737 | 1.11 |
|
|
|
| chr15_-_16948863 | 1.09 |
ENSDART00000062135
|
ENSDARG00000042379
|
ENSDARG00000042379 |
| chr6_-_39767452 | 1.06 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr3_-_48865474 | 1.04 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr23_-_9924987 | 1.01 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr8_+_8006488 | 1.01 |
|
|
|
| chr11_+_11217547 | 0.99 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr7_-_23775835 | 0.97 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr5_+_36487425 | 0.97 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr10_+_41746636 | 0.96 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr16_-_35633520 | 0.93 |
ENSDART00000169868
ENSDART00000166606 |
scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr9_-_41982635 | 0.93 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr15_-_16271208 | 0.92 |
|
|
|
| chr3_+_29891683 | 0.91 |
|
|
|
| chr7_+_25642061 | 0.89 |
ENSDART00000135728
|
hmgb3a
|
high mobility group box 3a |
| chr6_-_10086031 | 0.88 |
|
|
|
| chr14_+_33542705 | 0.85 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr21_+_23916512 | 0.84 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr14_+_13919450 | 0.83 |
ENSDART00000174760
|
nlgn3a
|
neuroligin 3a |
| chr4_+_25296408 | 0.82 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr3_+_23546802 | 0.80 |
ENSDART00000023674
|
hoxb9a
|
homeobox B9a |
| chr7_+_25641990 | 0.79 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr6_-_29297541 | 0.78 |
ENSDART00000078630
|
nme7
|
NME/NM23 family member 7 |
| chr20_-_10133201 | 0.77 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr13_+_22350043 | 0.75 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr7_+_20272091 | 0.74 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr12_-_44867369 | 0.74 |
ENSDART00000154859
|
trim65
|
tripartite motif containing 65 |
| chr4_+_25296376 | 0.72 |
ENSDART00000174536
|
sfmbt2
|
Scm-like with four mbt domains 2 |
| chr3_-_22082170 | 0.71 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr12_+_36716775 | 0.70 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr8_+_10267246 | 0.70 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr24_-_24010503 | 0.70 |
|
|
|
| chr16_+_25425398 | 0.69 |
|
|
|
| chr14_-_49115338 | 0.69 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr17_+_7438152 | 0.68 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr7_-_39281312 | 0.67 |
ENSDART00000170900
|
slc22a18
|
solute carrier family 22, member 18 |
| chr5_+_61926351 | 0.66 |
ENSDART00000132054
|
si:ch211-253h3.1
|
si:ch211-253h3.1 |
| chr6_-_35755435 | 0.64 |
ENSDART00000111642
|
brinp3a.1
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 1 |
| chr23_+_35964754 | 0.64 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr20_-_45819128 | 0.61 |
ENSDART00000124283
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr17_+_11264847 | 0.60 |
ENSDART00000153602
|
CU076099.1
|
ENSDARG00000097666 |
| chr17_+_443558 | 0.60 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr8_+_48488009 | 0.60 |
|
|
|
| chr5_+_37861949 | 0.59 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr9_-_1969197 | 0.58 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr3_-_23580055 | 0.58 |
|
|
|
| chr16_+_12101666 | 0.58 |
ENSDART00000143442
|
p3h3
|
prolyl 3-hydroxylase 3 |
| chr11_+_5745644 | 0.58 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr3_+_23557320 | 0.57 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr23_+_26215871 | 0.56 |
ENSDART00000158878
|
ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr14_+_35906366 | 0.56 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr6_+_29297594 | 0.54 |
ENSDART00000112099
|
zgc:172121
|
zgc:172121 |
| chr17_-_17744885 | 0.53 |
ENSDART00000155261
|
slirp
|
SRA stem-loop interacting RNA binding protein |
| chr17_-_45750693 | 0.53 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr10_-_20567013 | 0.53 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr19_+_34582100 | 0.51 |
ENSDART00000135592
|
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr24_+_30852751 | 0.51 |
|
|
|
| chr2_-_3203802 | 0.50 |
ENSDART00000105818
|
guk1a
|
guanylate kinase 1a |
| chr24_+_4946079 | 0.49 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr19_-_5416317 | 0.48 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr1_-_19900799 | 0.48 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr22_-_16971180 | 0.48 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr20_+_49273889 | 0.48 |
ENSDART00000166051
ENSDART00000112689 |
crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr5_+_26804344 | 0.47 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr4_+_12616987 | 0.46 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr12_+_27035744 | 0.46 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr24_-_36915249 | 0.43 |
ENSDART00000088130
|
dcakd
|
dephospho-CoA kinase domain containing |
| chr13_+_24132494 | 0.43 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr3_+_25247071 | 0.42 |
|
|
|
| chr19_+_32570656 | 0.42 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr4_+_12617165 | 0.42 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr18_+_9413668 | 0.42 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr2_+_37833161 | 0.41 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr24_-_5995073 | 0.41 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr1_+_18639896 | 0.41 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr25_+_6138600 | 0.40 |
ENSDART00000148995
|
slc25a44a
|
solute carrier family 25, member 44 a |
| chr11_-_38225034 | 0.39 |
ENSDART00000065613
|
elk4
|
ELK4, ETS-domain protein |
| chr2_+_27005381 | 0.39 |
|
|
|
| chr11_-_2133416 | 0.39 |
ENSDART00000170593
|
hoxc12b
|
homeobox C12b |
| chr22_-_16971035 | 0.38 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr19_+_14197118 | 0.38 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr12_-_31342432 | 0.38 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr3_+_23580216 | 0.38 |
ENSDART00000151584
|
hoxb4a
|
homeobox B4a |
| chr3_+_24059652 | 0.38 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr5_+_9542042 | 0.37 |
ENSDART00000137543
|
gstt2
|
glutathione S-transferase theta 2 |
| chr8_-_44186353 | 0.37 |
|
|
|
| chr22_+_31866783 | 0.37 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr3_+_17913109 | 0.36 |
ENSDART00000112384
|
ankrd40
|
ankyrin repeat domain 40 |
| chr23_+_27141681 | 0.36 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr13_-_37001997 | 0.36 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr7_+_15619409 | 0.35 |
ENSDART00000144765
ENSDART00000145946 |
pax6b
|
paired box 6b |
| chr15_-_12338925 | 0.34 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr24_+_25326286 | 0.34 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr16_-_45321894 | 0.33 |
ENSDART00000044097
|
hpn
|
hepsin |
| chr2_-_57502941 | 0.33 |
|
|
|
| chr4_-_4923982 | 0.33 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr7_+_62010604 | 0.32 |
|
|
|
| chr12_-_29190576 | 0.31 |
ENSDART00000153458
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr21_+_37726304 | 0.31 |
ENSDART00000076320
|
mrps17
|
mitochondrial ribosomal protein S17 |
| chr4_+_25868802 | 0.31 |
|
|
|
| chr5_-_67043355 | 0.31 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr17_-_45750643 | 0.30 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr12_-_844493 | 0.30 |
ENSDART00000022688
|
tob1b
|
transducer of ERBB2, 1b |
| chr12_+_9779597 | 0.30 |
ENSDART00000137081
|
rundc3ab
|
RUN domain containing 3Ab |
| chr15_+_6870469 | 0.30 |
|
|
|
| chr1_-_29885008 | 0.29 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr22_-_16970979 | 0.28 |
ENSDART00000090237
|
nfia
|
nuclear factor I/A |
| chr22_-_20101177 | 0.28 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr7_+_62010636 | 0.28 |
|
|
|
| chr9_-_43340836 | 0.28 |
ENSDART00000059451
|
ccdc141
|
coiled-coil domain containing 141 |
| chr24_+_25326233 | 0.28 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr17_+_41475052 | 0.28 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr11_-_38225208 | 0.27 |
ENSDART00000102850
|
elk4
|
ELK4, ETS-domain protein |
| chr23_-_24562107 | 0.27 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr1_-_49274993 | 0.26 |
ENSDART00000050603
|
hadh
|
hydroxyacyl-CoA dehydrogenase |
| chr8_+_53241686 | 0.26 |
ENSDART00000125232
|
nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr2_-_30005139 | 0.26 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr7_-_18894589 | 0.25 |
ENSDART00000142924
|
kirrel1a
|
kin of IRRE like a |
| chr16_+_24057158 | 0.24 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr1_-_25225339 | 0.24 |
ENSDART00000109714
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr9_+_1968455 | 0.23 |
|
|
|
| chr22_-_17570275 | 0.23 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr2_-_23331500 | 0.23 |
ENSDART00000146497
|
mllt1b
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 1b |
| chr19_+_42492029 | 0.22 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr7_-_68417190 | 0.22 |
|
|
|
| chr2_-_8890701 | 0.21 |
ENSDART00000010257
|
fam73a
|
family with sequence similarity 73, member A |
| chr1_+_50348973 | 0.21 |
|
|
|
| chr17_+_7437994 | 0.21 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr6_-_50405426 | 0.19 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr6_-_45993454 | 0.18 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr8_+_10785326 | 0.17 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr16_-_30847364 | 0.17 |
|
|
|
| chr14_+_49056502 | 0.16 |
ENSDART00000124773
|
ppid
|
peptidylprolyl isomerase D |
| chr19_-_3547521 | 0.15 |
ENSDART00000168139
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr20_+_31381039 | 0.15 |
|
|
|
| chr21_-_36886505 | 0.15 |
ENSDART00000113678
|
wwc1
|
WW and C2 domain containing 1 |
| chr14_-_33468547 | 0.14 |
|
|
|
| chr25_+_21919595 | 0.14 |
ENSDART00000156517
|
ENSDARG00000062199
|
ENSDARG00000062199 |
| chr8_-_15955746 | 0.13 |
ENSDART00000132908
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr4_-_16461881 | 0.12 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr5_+_58824344 | 0.12 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr19_+_14197020 | 0.12 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr5_-_67043168 | 0.12 |
ENSDART00000134386
|
arhgap31
|
Rho GTPase activating protein 31 |
| chr1_-_51100156 | 0.12 |
ENSDART00000109640
|
junba
|
jun B proto-oncogene a |
| chr3_+_31047120 | 0.12 |
|
|
|
| chr12_+_44867426 | 0.12 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr9_+_24255064 | 0.12 |
ENSDART00000101577
ENSDART00000159324 ENSDART00000023196 ENSDART00000079689 ENSDART00000172743 ENSDART00000171577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr13_-_37002066 | 0.11 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr4_-_18786577 | 0.10 |
ENSDART00000141187
|
def6c
|
differentially expressed in FDCP 6c homolog |
| chr6_+_12972739 | 0.10 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr24_-_21027589 | 0.09 |
ENSDART00000154259
|
atp6v1ab
|
ATPase, H+ transporting, lysosomal, V1 subunit Ab |
| chr8_-_16472564 | 0.09 |
|
|
|
| chr23_-_35976816 | 0.09 |
|
|
|
| chr17_+_7438085 | 0.09 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr8_-_14014576 | 0.09 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr4_-_17033790 | 0.08 |
|
|
|
| chr7_-_20565207 | 0.08 |
ENSDART00000052977
|
cldn15a
|
claudin 15a |
| chr5_-_67414115 | 0.08 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr5_-_18990190 | 0.08 |
ENSDART00000098795
|
mmab
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr24_-_39313011 | 0.07 |
|
|
|
| chr6_+_29799599 | 0.07 |
|
|
|
| chr19_+_17898639 | 0.07 |
|
|
|
| chr8_+_13069201 | 0.07 |
ENSDART00000138433
|
itgb4
|
integrin, beta 4 |
| chr15_+_28335293 | 0.06 |
ENSDART00000152536
|
myo1cb
|
myosin Ic, paralog b |
| chr13_+_24132236 | 0.06 |
ENSDART00000088005
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr12_+_9779839 | 0.06 |
ENSDART00000123712
|
rundc3ab
|
RUN domain containing 3Ab |
| chr12_+_36716838 | 0.06 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr9_+_41910191 | 0.06 |
|
|
|
| chr18_+_18465885 | 0.06 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr14_-_30811243 | 0.05 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr5_-_16479187 | 0.05 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr24_-_16741851 | 0.05 |
|
|
|
| chr11_+_5745962 | 0.04 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr3_+_32711007 | 0.04 |
ENSDART00000122228
|
prr14
|
proline rich 14 |
| chr24_+_2962301 | 0.03 |
ENSDART00000170835
|
eci2
|
enoyl-CoA delta isomerase 2 |
| chr22_+_39067994 | 0.03 |
|
|
|
| chr13_+_18376642 | 0.02 |
ENSDART00000142622
|
si:ch211-198a12.6
|
si:ch211-198a12.6 |
| chr13_+_31473510 | 0.02 |
ENSDART00000030646
|
six6a
|
SIX homeobox 6a |
| chr10_-_30006277 | 0.01 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr20_-_27038165 | 0.00 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.6 | 1.7 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.5 | 2.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.3 | 0.8 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 2.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.4 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.5 | GO:1902369 | negative regulation of RNA catabolic process(GO:1902369) |
| 0.1 | 0.4 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 2.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.7 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 0.5 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.5 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.1 | 0.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.4 | GO:0090104 | pancreatic epsilon cell differentiation(GO:0090104) |
| 0.1 | 0.4 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.8 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.4 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.6 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.8 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.5 | GO:0009086 | methionine metabolic process(GO:0006555) methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.7 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 2.8 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.6 | GO:0044259 | collagen metabolic process(GO:0032963) multicellular organismal macromolecule metabolic process(GO:0044259) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.0 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.6 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 1.1 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0015740 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) C4-dicarboxylate transport(GO:0015740) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.1 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 0.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 3.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.5 | GO:0000974 | Prp19 complex(GO:0000974) U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 1.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 1.7 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 2.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 1.4 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.2 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.6 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.4 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.5 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.1 | 0.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 1.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.6 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.9 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 2.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 3.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 3.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.4 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |