Project
DANIO-CODE
Navigation
Downloads
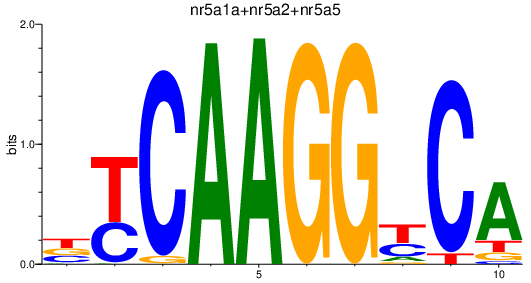
Results for nr5a1a+nr5a2+nr5a5
Z-value: 0.92
Transcription factors associated with nr5a1a+nr5a2+nr5a5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr5a5
|
ENSDARG00000039116 | nuclear receptor subfamily 5, group A, member 5 |
|
nr5a2
|
ENSDARG00000100940 | nuclear receptor subfamily 5, group A, member 2 |
|
nr5a1a
|
ENSDARG00000103176 | nuclear receptor subfamily 5, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr5a2 | dr10_dc_chr22_-_22694364_22694419 | 0.79 | 2.6e-04 | Click! |
Activity profile of nr5a1a+nr5a2+nr5a5 motif
Sorted Z-values of nr5a1a+nr5a2+nr5a5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr5a1a+nr5a2+nr5a5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_30816780 | 2.76 |
ENSDART00000103475
|
agr2
|
anterior gradient 2 |
| chr2_+_42342148 | 1.95 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr7_-_25623974 | 1.94 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr9_-_1969197 | 1.94 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr23_+_25782195 | 1.81 |
ENSDART00000060059
|
rbms2b
|
RNA binding motif, single stranded interacting protein 2b |
| chr7_-_28425307 | 1.78 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr3_+_1413640 | 1.77 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr20_+_51388214 | 1.77 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr14_-_31676235 | 1.73 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
| chr13_-_13163676 | 1.73 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr5_+_55598181 | 1.69 |
ENSDART00000021159
|
lhx1b
|
LIM homeobox 1b |
| chr22_+_33177432 | 1.69 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr7_+_25642061 | 1.61 |
ENSDART00000135728
|
hmgb3a
|
high mobility group box 3a |
| chr22_-_2870591 | 1.61 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr20_+_230394 | 1.54 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr19_-_12226198 | 1.54 |
ENSDART00000032474
ENSDART00000151056 ENSDART00000167360 |
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr6_-_15526547 | 1.49 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr20_+_51388313 | 1.46 |
ENSDART00000153452
|
hsp90ab1
|
heat shock protein 90, alpha (cytosolic), class B member 1 |
| chr7_-_8635687 | 1.44 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr19_+_5375413 | 1.44 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr25_-_4376129 | 1.43 |
|
|
|
| chr4_-_21931540 | 1.42 |
ENSDART00000174400
|
rps16
|
ribosomal protein S16 |
| chr7_+_25641990 | 1.41 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr23_+_17856053 | 1.36 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr7_-_57941497 | 1.36 |
ENSDART00000114008
|
unm_hu7910
|
un-named hu7910 |
| chr11_-_7310284 | 1.35 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr13_-_22877318 | 1.33 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr3_+_27684309 | 1.33 |
|
|
|
| chr5_+_26829686 | 1.32 |
ENSDART00000087894
|
zgc:153409
|
zgc:153409 |
| chr5_-_21523520 | 1.27 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr3_-_26393945 | 1.25 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr2_-_20941256 | 1.25 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr7_+_34417030 | 1.24 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr14_+_34174018 | 1.24 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr18_+_20504980 | 1.23 |
ENSDART00000060295
|
rapsn
|
receptor-associated protein of the synapse, 43kD |
| chr2_-_28244864 | 1.20 |
ENSDART00000137542
|
zgc:163121
|
zgc:163121 |
| chr5_-_34701639 | 1.20 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr12_-_48493654 | 1.20 |
ENSDART00000162603
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr24_-_16884946 | 1.19 |
ENSDART00000171988
|
ptgdsb.2
|
prostaglandin D2 synthase b, tandem duplicate 2 |
| chr7_-_33080261 | 1.13 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr7_-_23933499 | 1.13 |
ENSDART00000087298
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr19_+_14197118 | 1.12 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr13_-_13163801 | 1.12 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr2_+_30932612 | 1.11 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr15_-_26619404 | 1.10 |
ENSDART00000150152
|
serpinf2b
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2b |
| chr7_+_20215723 | 1.10 |
ENSDART00000173724
ENSDART00000173773 |
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr7_-_25816549 | 1.09 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr12_-_457997 | 1.08 |
ENSDART00000143232
|
dhrs7cb
|
dehydrogenase/reductase (SDR family) member 7Cb |
| chr3_-_39287733 | 1.07 |
|
|
|
| chr18_-_25010085 | 1.06 |
ENSDART00000133786
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr7_-_25816655 | 1.03 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr19_+_21140010 | 1.02 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr15_+_37204488 | 1.01 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr16_-_21120115 | 1.01 |
|
|
|
| chr23_+_25428372 | 0.99 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr20_+_34417665 | 0.98 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr3_-_21149752 | 0.98 |
ENSDART00000003939
|
syngr1a
|
synaptogyrin 1a |
| chr4_+_14361566 | 0.98 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr5_-_70939686 | 0.98 |
|
|
|
| chr5_-_34701743 | 0.97 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr16_+_32605735 | 0.97 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr2_-_58272475 | 0.97 |
ENSDART00000166282
ENSDART00000159040 |
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr3_-_32727588 | 0.96 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr14_-_1081316 | 0.96 |
|
|
|
| chr19_-_9793494 | 0.95 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr6_-_39346614 | 0.95 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr16_+_55351732 | 0.95 |
ENSDART00000109391
|
CABZ01052290.1
|
ENSDARG00000077559 |
| chr14_-_5371581 | 0.95 |
ENSDART00000012116
|
tlx2
|
T-cell leukemia, homeobox 2 |
| chr14_+_5852295 | 0.94 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr1_-_4757890 | 0.94 |
ENSDART00000114035
|
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr18_+_17594381 | 0.93 |
ENSDART00000010998
|
slc12a3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr3_-_40159349 | 0.93 |
ENSDART00000055186
|
atp5j2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 |
| chr18_-_15963983 | 0.92 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr8_+_10267246 | 0.92 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr14_-_11947241 | 0.91 |
ENSDART00000115101
|
myot
|
myotilin |
| chr4_-_16356071 | 0.91 |
ENSDART00000079521
|
kera
|
keratocan |
| chr9_+_30774173 | 0.90 |
ENSDART00000160590
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr23_+_18796386 | 0.89 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr19_+_47719438 | 0.88 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr25_-_34599079 | 0.87 |
ENSDART00000156727
|
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr6_+_20276828 | 0.87 |
ENSDART00000128047
|
uqcrc1
|
ubiquinol-cytochrome c reductase core protein I |
| chr25_-_21718782 | 0.87 |
ENSDART00000175132
|
ENSDARG00000051873
|
ENSDARG00000051873 |
| chr2_+_43736542 | 0.87 |
ENSDART00000148454
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr5_+_50947650 | 0.86 |
ENSDART00000058403
|
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr8_-_11806627 | 0.86 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr11_+_30416115 | 0.86 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
| chr15_-_1225595 | 0.86 |
|
|
|
| chr16_+_53221957 | 0.84 |
|
|
|
| chr22_-_10324166 | 0.84 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr4_+_22754618 | 0.84 |
|
|
|
| chr8_-_52729505 | 0.84 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr7_-_58540731 | 0.83 |
ENSDART00000167231
|
mrpl15
|
mitochondrial ribosomal protein L15 |
| chr20_-_28981879 | 0.81 |
ENSDART00000134564
ENSDART00000132127 |
srsf5b
|
serine/arginine-rich splicing factor 5b |
| chr20_+_230164 | 0.81 |
ENSDART00000002661
|
lama4
|
laminin, alpha 4 |
| chr3_-_52419408 | 0.80 |
ENSDART00000154260
|
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr6_+_41557636 | 0.80 |
ENSDART00000084834
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr2_+_37985514 | 0.80 |
ENSDART00000149224
|
apol1
|
apolipoprotein L, 1 |
| chr19_-_27807312 | 0.80 |
|
|
|
| chr12_-_3903961 | 0.80 |
ENSDART00000134292
|
ENSDARG00000021154
|
ENSDARG00000021154 |
| chr20_+_7337738 | 0.79 |
ENSDART00000165596
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr23_+_17856104 | 0.79 |
ENSDART00000154427
|
CR381647.1
|
ENSDARG00000097211 |
| chr8_-_23102555 | 0.78 |
|
|
|
| chr10_+_29351676 | 0.78 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr13_-_2083873 | 0.77 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr21_-_19789339 | 0.77 |
|
|
|
| chr1_+_9999842 | 0.77 |
ENSDART00000152438
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr7_-_57727148 | 0.77 |
|
|
|
| chr9_-_28292714 | 0.76 |
ENSDART00000146284
|
kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr5_-_24568752 | 0.76 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr24_+_26287471 | 0.75 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr25_+_1747864 | 0.75 |
|
|
|
| chr8_+_23192085 | 0.75 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr10_+_37983342 | 0.75 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr23_+_25930072 | 0.74 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr17_+_7438152 | 0.72 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr4_-_72097569 | 0.72 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr17_-_12231165 | 0.71 |
ENSDART00000080927
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr23_+_14058972 | 0.71 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr12_-_36415239 | 0.70 |
|
|
|
| chr20_+_26408687 | 0.70 |
|
|
|
| chr12_-_25110150 | 0.70 |
ENSDART00000077188
|
cox7a3
|
cytochrome c oxidase subunit VIIa polypeptide 3 |
| chr18_+_41323776 | 0.70 |
ENSDART00000048985
|
veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr23_-_19053587 | 0.69 |
|
|
|
| chr10_+_9259213 | 0.68 |
ENSDART00000064966
|
snx18b
|
sorting nexin 18b |
| chr16_-_30698002 | 0.68 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr17_-_26592356 | 0.68 |
ENSDART00000016608
|
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr12_+_7421078 | 0.67 |
ENSDART00000163114
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr19_-_9793444 | 0.67 |
ENSDART00000134816
|
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr21_-_20305406 | 0.67 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr11_-_18442094 | 0.67 |
ENSDART00000159380
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr15_-_4536026 | 0.67 |
|
|
|
| chr23_+_35988395 | 0.66 |
ENSDART00000154825
|
hoxc3a
|
homeo box C3a |
| chr7_-_71958596 | 0.66 |
|
|
|
| chr14_-_24814149 | 0.66 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr7_-_8635989 | 0.65 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr10_-_17383380 | 0.65 |
|
|
|
| chr19_+_10912601 | 0.65 |
ENSDART00000053325
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr19_-_27878747 | 0.64 |
ENSDART00000137638
|
si:ch73-25f10.6
|
si:ch73-25f10.6 |
| chr8_+_7237516 | 0.64 |
|
|
|
| chr16_-_22903823 | 0.63 |
|
|
|
| chr22_+_31866783 | 0.63 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr13_+_50062513 | 0.61 |
ENSDART00000124142
ENSDART00000099537 |
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr13_-_18064929 | 0.61 |
ENSDART00000079902
|
slc25a16
|
solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 |
| chr7_-_27877036 | 0.61 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr14_-_29487265 | 0.61 |
|
|
|
| chr17_-_50899135 | 0.57 |
|
|
|
| chr2_-_2509717 | 0.56 |
ENSDART00000082143
ENSDART00000158864 |
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr11_-_29908609 | 0.56 |
ENSDART00000156196
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr12_-_34621359 | 0.56 |
|
|
|
| KN149817v1_+_2207 | 0.55 |
|
|
|
| chr22_+_12406830 | 0.55 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr13_+_41998500 | 0.54 |
ENSDART00000074707
|
cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3 |
| chr2_+_32796877 | 0.54 |
ENSDART00000082250
|
zgc:136930
|
zgc:136930 |
| chr23_-_22186674 | 0.53 |
ENSDART00000142474
|
phc2a
|
polyhomeotic homolog 2a (Drosophila) |
| chr10_-_32580373 | 0.53 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr9_+_17341042 | 0.53 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr25_-_35094733 | 0.53 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr1_-_44239584 | 0.53 |
ENSDART00000137216
|
tmem176
|
transmembrane protein 176 |
| chr4_+_25637474 | 0.52 |
ENSDART00000066942
ENSDART00000041965 |
acot15
|
acyl-CoA thioesterase 15 |
| chr3_+_19863136 | 0.52 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr11_+_2786083 | 0.52 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr20_+_35183329 | 0.52 |
ENSDART00000114262
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr14_-_35074225 | 0.51 |
ENSDART00000129460
|
rnaseh2c
|
ribonuclease H2, subunit C |
| chr18_-_47186938 | 0.51 |
|
|
|
| chr3_-_39287813 | 0.50 |
|
|
|
| chr13_-_29290894 | 0.50 |
ENSDART00000150228
|
chata
|
choline O-acetyltransferase a |
| chr5_-_31845561 | 0.49 |
ENSDART00000157295
ENSDART00000159522 |
ncs1a
|
neuronal calcium sensor 1a |
| chr10_-_17383466 | 0.49 |
|
|
|
| chr10_+_42546351 | 0.49 |
ENSDART00000020000
|
COX5B
|
cytochrome c oxidase subunit 5B |
| chr20_-_53162473 | 0.48 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr17_-_31041977 | 0.48 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr2_+_289138 | 0.48 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr17_+_21433543 | 0.47 |
ENSDART00000079030
|
pla2g4f.1
|
phospholipase A2, group IVF, tandem duplicate 1 |
| chr9_+_33334773 | 0.47 |
ENSDART00000005879
|
atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr7_-_25624212 | 0.47 |
ENSDART00000173505
|
cd99l2
|
CD99 molecule-like 2 |
| chr7_-_25816498 | 0.47 |
ENSDART00000052989
ENSDART00000047951 |
ache
|
acetylcholinesterase |
| chr15_+_26156180 | 0.46 |
|
|
|
| chr14_+_38505346 | 0.46 |
ENSDART00000129958
ENSDART00000144599 |
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr18_-_48498261 | 0.46 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr1_+_44239727 | 0.45 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr6_+_29716538 | 0.45 |
ENSDART00000164849
|
phb2b
|
prohibitin 2b |
| chr20_-_25839801 | 0.44 |
|
|
|
| chr19_+_14197020 | 0.44 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr15_-_25430003 | 0.44 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr17_+_23102259 | 0.44 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr11_-_2563343 | 0.43 |
ENSDART00000008133
|
ENSDARG00000002897
|
ENSDARG00000002897 |
| chr24_+_24309660 | 0.43 |
|
|
|
| chr23_+_35984675 | 0.43 |
ENSDART00000053295
|
hoxc10a
|
homeobox C10a |
| chr23_-_20026717 | 0.43 |
ENSDART00000153828
|
atp2b3b
|
ATPase, Ca++ transporting, plasma membrane 3b |
| chr18_+_20504641 | 0.42 |
|
|
|
| chr25_-_21718647 | 0.41 |
ENSDART00000152014
ENSDART00000177876 |
ENSDARG00000051873
|
ENSDARG00000051873 |
| chr14_-_1081393 | 0.41 |
|
|
|
| chr9_+_21908880 | 0.41 |
|
|
|
| chr16_-_5821719 | 0.40 |
ENSDART00000136655
|
ndufa3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 |
| chr17_-_43296680 | 0.40 |
ENSDART00000176637
|
EML5
|
echinoderm microtubule associated protein like 5 |
| chr16_+_14135089 | 0.40 |
|
|
|
| chr3_+_60724858 | 0.40 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr10_-_7712439 | 0.38 |
ENSDART00000159330
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr17_-_33336706 | 0.38 |
ENSDART00000040346
|
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr3_-_8608620 | 0.38 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0014075 | response to amphetamine(GO:0001975) acetylcholine catabolic process(GO:0006581) response to amine(GO:0014075) |
| 0.8 | 3.2 | GO:1901388 | transforming growth factor beta activation(GO:0036363) transforming growth factor beta production(GO:0071604) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) regulation of transforming growth factor beta activation(GO:1901388) negative regulation of transforming growth factor beta activation(GO:1901389) |
| 0.6 | 1.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.5 | 2.8 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.5 | 1.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 1.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 1.0 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.3 | 4.0 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.3 | 1.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 0.5 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 3.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 0.8 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 2.1 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.2 | 0.5 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:0060394 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.9 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.3 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 0.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.4 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 3.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.4 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 0.4 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.9 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.9 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 2.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 1.3 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.1 | 0.9 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 1.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.8 | GO:0035775 | pronephric glomerulus morphogenesis(GO:0035775) |
| 0.1 | 0.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.9 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 1.4 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 1.0 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 1.2 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.6 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.8 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 1.0 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.9 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.8 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 1.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.3 | GO:0009081 | branched-chain amino acid metabolic process(GO:0009081) |
| 0.0 | 1.1 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.7 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 2.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 3.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 1.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 1.8 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.3 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 2.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.6 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 2.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0003990 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.4 | 1.3 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.3 | 1.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 0.9 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 1.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.2 | 1.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 1.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.2 | 3.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 3.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 1.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.9 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.7 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.4 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 0.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 1.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.9 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.3 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.4 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.9 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.5 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.9 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.3 | GO:0004691 | AMP-activated protein kinase activity(GO:0004679) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 3.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.6 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 1.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 2.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 3.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 3.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 0.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.9 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 2.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 3.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.5 | 3.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 2.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 1.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.6 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.4 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |