Project
DANIO-CODE
Navigation
Downloads
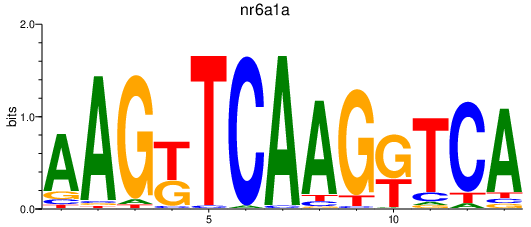
Results for nr6a1a
Z-value: 1.14
Transcription factors associated with nr6a1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1a
|
ENSDARG00000101508 | nuclear receptor subfamily 6, group A, member 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1a | dr10_dc_chr8_-_52952226_52952290 | -0.72 | 1.5e-03 | Click! |
Activity profile of nr6a1a motif
Sorted Z-values of nr6a1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_36503296 | 3.17 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr5_-_11471707 | 1.97 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr25_+_7318688 | 1.89 |
ENSDART00000166496
|
cat
|
catalase |
| chr17_-_45021393 | 1.83 |
|
|
|
| chr9_-_12687806 | 1.64 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr10_+_33449922 | 1.58 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr8_-_22265875 | 1.55 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr18_+_8954323 | 1.54 |
ENSDART00000134827
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr7_+_15781933 | 1.44 |
ENSDART00000161669
|
immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr19_-_5186692 | 1.39 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr1_+_23866532 | 1.39 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr24_-_9844547 | 1.31 |
|
|
|
| chr12_+_30673985 | 1.30 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_+_22076136 | 1.28 |
ENSDART00000080877
|
tnfsf10l4
|
tumor necrosis factor (ligand) superfamily, member 10 like 4 |
| chr11_+_44909949 | 1.24 |
ENSDART00000173116
ENSDART00000167011 ENSDART00000167226 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr2_-_50491234 | 1.19 |
ENSDART00000165678
|
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr2_-_22631167 | 1.13 |
ENSDART00000168653
ENSDART00000158558 |
fam110b
BX927330.1
|
family with sequence similarity 110, member B ENSDARG00000098356 |
| chr14_+_35074248 | 1.07 |
ENSDART00000084914
|
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr21_-_804453 | 1.05 |
|
|
|
| chr8_-_26690715 | 1.01 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr1_+_49170967 | 1.00 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr5_-_36503236 | 0.99 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr24_+_34183462 | 0.99 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr7_+_24855467 | 0.98 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr8_-_22266041 | 0.97 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr1_-_7882397 | 0.96 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr10_-_36849540 | 0.91 |
|
|
|
| chr3_-_49031058 | 0.90 |
ENSDART00000154561
|
mrpl12
|
mitochondrial ribosomal protein L12 |
| chr18_-_20619155 | 0.90 |
ENSDART00000151864
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr11_+_24076576 | 0.89 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr3_-_58400454 | 0.89 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr10_-_36849596 | 0.89 |
|
|
|
| chr7_-_31346991 | 0.89 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr8_+_15213841 | 0.88 |
ENSDART00000063717
|
ENSDARG00000043402
|
ENSDARG00000043402 |
| chr14_-_33138710 | 0.87 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr24_-_9838947 | 0.86 |
ENSDART00000138576
|
zgc:171977
|
zgc:171977 |
| chr11_+_26895952 | 0.86 |
ENSDART00000158411
ENSDART00000173374 ENSDART00000129736 |
hdac11
|
histone deacetylase 11 |
| chr15_-_16137131 | 0.85 |
|
|
|
| chr11_-_7786415 | 0.84 |
ENSDART00000154569
|
CR450813.1
|
ENSDARG00000097035 |
| chr8_+_26838596 | 0.83 |
|
|
|
| chr24_+_34183557 | 0.83 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr20_+_28006043 | 0.83 |
|
|
|
| chr15_+_30246345 | 0.80 |
ENSDART00000100214
|
nufip2
|
nuclear fragile X mental retardation protein interacting protein 2 |
| chr13_-_23825684 | 0.79 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr11_-_18637482 | 0.78 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr7_-_31346844 | 0.78 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr21_-_32341308 | 0.78 |
ENSDART00000112550
|
mapk9
|
mitogen-activated protein kinase 9 |
| chr14_+_25927045 | 0.77 |
ENSDART00000168673
ENSDART00000128971 |
gm2a
|
GM2 ganglioside activator |
| chr13_-_11630642 | 0.77 |
ENSDART00000102381
|
ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_29733037 | 0.76 |
ENSDART00000133955
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr15_-_16950049 | 0.76 |
|
|
|
| chr7_-_25252260 | 0.74 |
ENSDART00000173657
|
CU929416.1
|
ENSDARG00000105572 |
| chr6_+_42340776 | 0.73 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr13_-_8360779 | 0.72 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr10_-_16910230 | 0.72 |
ENSDART00000171755
ENSDART00000170953 |
stoml2
|
stomatin (EPB72)-like 2 |
| chr16_-_19155418 | 0.71 |
ENSDART00000134796
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr24_+_17190448 | 0.71 |
ENSDART00000159724
ENSDART00000081851 ENSDART00000018868 |
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr18_-_21199617 | 0.71 |
ENSDART00000060166
|
got2a
|
glutamic-oxaloacetic transaminase 2a, mitochondrial |
| chr2_+_38959335 | 0.70 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr7_-_31346802 | 0.70 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr22_+_1643493 | 0.69 |
ENSDART00000167767
|
BX649416.1
|
ENSDARG00000098230 |
| chr5_+_61711614 | 0.69 |
ENSDART00000082965
|
ABR
|
active BCR-related |
| chr1_-_46260853 | 0.67 |
|
|
|
| chr17_-_42523585 | 0.67 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr14_+_29932533 | 0.67 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr12_-_4597113 | 0.66 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr16_+_29574449 | 0.66 |
ENSDART00000148450
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr8_-_26690680 | 0.66 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr13_-_11630475 | 0.66 |
ENSDART00000035505
|
ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_12087549 | 0.65 |
|
|
|
| chr19_+_24334831 | 0.65 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr1_-_7968497 | 0.65 |
ENSDART00000141407
ENSDART00000148067 |
actb1
|
actin, beta 1 |
| chr22_-_6723094 | 0.64 |
|
|
|
| chr25_+_7318825 | 0.64 |
ENSDART00000159748
|
cat
|
catalase |
| chr10_+_43289261 | 0.64 |
ENSDART00000024644
|
xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr16_-_17289585 | 0.64 |
ENSDART00000135146
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr9_+_8919663 | 0.63 |
ENSDART00000134954
|
carkd
|
carbohydrate kinase domain containing |
| chr4_-_71644701 | 0.62 |
ENSDART00000174178
|
si:cabz01021430.2
|
si:cabz01021430.2 |
| chr4_+_8007952 | 0.62 |
ENSDART00000014036
|
optn
|
optineurin |
| chr25_+_16796766 | 0.61 |
|
|
|
| chr23_-_27647386 | 0.61 |
|
|
|
| chr17_-_42523635 | 0.61 |
ENSDART00000154755
|
prkd3
|
protein kinase D3 |
| chr17_+_18217416 | 0.61 |
ENSDART00000156616
|
BX465857.1
|
ENSDARG00000097921 |
| chr8_-_11806733 | 0.60 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr10_-_36849472 | 0.60 |
|
|
|
| chr8_-_22265802 | 0.60 |
ENSDART00000134033
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr17_+_5680342 | 0.60 |
ENSDART00000156089
|
CU929322.1
|
ENSDARG00000096839 |
| chr5_-_40524177 | 0.59 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr14_-_33114322 | 0.58 |
ENSDART00000173267
|
tmem255a
|
transmembrane protein 255A |
| chr9_-_10833577 | 0.57 |
ENSDART00000078348
ENSDART00000134911 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr15_+_28477741 | 0.57 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr6_-_7529384 | 0.57 |
ENSDART00000091836
ENSDART00000151697 |
ubn2a
|
ubinuclein 2a |
| chr10_-_42302932 | 0.57 |
ENSDART00000076693
ENSDART00000073631 |
stambpa
|
STAM binding protein a |
| chr3_-_58400345 | 0.57 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr1_-_42915971 | 0.57 |
|
|
|
| chr18_+_6747156 | 0.56 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr14_-_21320696 | 0.56 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr11_+_26125813 | 0.56 |
ENSDART00000088800
|
ppcs
|
phosphopantothenoylcysteine synthetase |
| chr7_-_7533176 | 0.55 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr13_+_42183685 | 0.55 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr7_+_55648832 | 0.55 |
ENSDART00000082780
|
ACSF3
|
acyl-CoA synthetase family member 3 |
| chr19_-_6503643 | 0.55 |
|
|
|
| chr13_-_8979137 | 0.54 |
ENSDART00000058056
|
mrps26
|
mitochondrial ribosomal protein S26 |
| chr8_+_13757180 | 0.54 |
|
|
|
| chr8_-_54103635 | 0.53 |
|
|
|
| chr23_+_25808393 | 0.53 |
|
|
|
| chr5_+_40599287 | 0.52 |
ENSDART00000137545
|
BX005419.1
|
ENSDARG00000093616 |
| chr10_+_33450122 | 0.52 |
ENSDART00000115379
ENSDART00000163458 ENSDART00000078012 |
zgc:153345
|
zgc:153345 |
| chr24_+_24999138 | 0.52 |
ENSDART00000152095
ENSDART00000154988 ENSDART00000156098 |
CR383672.2
|
ENSDARG00000096452 |
| chr11_-_18637646 | 0.52 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr2_-_59019569 | 0.51 |
|
|
|
| chr7_+_7261692 | 0.51 |
ENSDART00000102629
|
nit1
|
nitrilase 1 |
| chr13_+_50167246 | 0.51 |
ENSDART00000178572
|
CU570781.2
|
ENSDARG00000108623 |
| chr13_+_25722545 | 0.50 |
|
|
|
| chr10_-_36849734 | 0.50 |
|
|
|
| chr8_+_13757024 | 0.50 |
|
|
|
| chr5_-_54578261 | 0.50 |
ENSDART00000016995
ENSDART00000134937 |
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr11_+_26896056 | 0.49 |
ENSDART00000158411
ENSDART00000173374 ENSDART00000129736 |
hdac11
|
histone deacetylase 11 |
| chr4_+_16020464 | 0.49 |
ENSDART00000144611
|
CR749763.5
|
ENSDARG00000093983 |
| chr12_-_18286666 | 0.49 |
ENSDART00000078767
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr22_+_25059272 | 0.48 |
ENSDART00000177284
|
rrbp1b
|
ribosome binding protein 1b |
| chr9_-_21427035 | 0.48 |
ENSDART00000146764
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr14_-_36072158 | 0.48 |
ENSDART00000128244
|
asb5a
|
ankyrin repeat and SOCS box containing 5a |
| chr21_+_15617110 | 0.47 |
ENSDART00000015841
|
gstt1b
|
glutathione S-transferase theta 1b |
| chr5_-_15921657 | 0.47 |
|
|
|
| chr1_+_16834347 | 0.47 |
ENSDART00000005593
ENSDART00000140076 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr2_+_58448918 | 0.46 |
ENSDART00000098173
|
vapal
|
VAMP (vesicle-associated membrane protein)-associated protein A, like |
| chr14_+_35074075 | 0.46 |
ENSDART00000084914
|
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr9_-_10833656 | 0.46 |
ENSDART00000078348
ENSDART00000134911 |
si:ch1073-416j23.1
|
si:ch1073-416j23.1 |
| chr12_+_33294613 | 0.46 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr17_+_18097490 | 0.45 |
ENSDART00000144894
|
bcl11ba
|
B-cell CLL/lymphoma 11Ba (zinc finger protein) |
| chr5_-_47521160 | 0.45 |
ENSDART00000144252
|
BX465834.1
|
ENSDARG00000095715 |
| chr17_+_28532738 | 0.44 |
|
|
|
| chr9_+_24255311 | 0.44 |
ENSDART00000166303
|
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr9_-_22377136 | 0.44 |
ENSDART00000110933
|
CR847505.1
|
ENSDARG00000076693 |
| chr15_-_31104074 | 0.44 |
ENSDART00000157005
|
nf1a
|
neurofibromin 1a |
| chr7_-_67508752 | 0.44 |
ENSDART00000167891
|
CR388184.1
|
ENSDARG00000099896 |
| chr21_-_21142020 | 0.44 |
ENSDART00000079678
|
ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr12_+_11314303 | 0.44 |
ENSDART00000129495
|
si:rp71-19m20.1
|
si:rp71-19m20.1 |
| chr6_-_11127409 | 0.44 |
ENSDART00000151125
|
pcnt
|
pericentrin |
| chr14_+_15728850 | 0.43 |
ENSDART00000161348
|
prelid1a
|
PRELI domain containing 1a |
| chr7_+_61450760 | 0.43 |
ENSDART00000165864
|
acox3
|
acyl-CoA oxidase 3, pristanoyl |
| chr25_-_28636834 | 0.42 |
ENSDART00000138087
|
cox5aa
|
cytochrome c oxidase subunit Vaa |
| chr15_-_30247084 | 0.42 |
ENSDART00000154069
|
BX322555.4
|
ENSDARG00000097908 |
| chr9_-_21427331 | 0.42 |
ENSDART00000102143
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr5_-_54578223 | 0.42 |
ENSDART00000016995
ENSDART00000134937 |
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr24_-_10253650 | 0.41 |
ENSDART00000127568
ENSDART00000106260 |
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr2_+_25212281 | 0.41 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr24_+_17190237 | 0.41 |
ENSDART00000126435
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr12_-_18286637 | 0.40 |
ENSDART00000078767
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr18_-_2954175 | 0.39 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr25_-_34638406 | 0.38 |
ENSDART00000123853
|
hist1h4l
|
histone 1, H4, like |
| chr13_+_50167216 | 0.38 |
ENSDART00000178572
|
CU570781.2
|
ENSDARG00000108623 |
| chr10_-_16910257 | 0.38 |
ENSDART00000171755
ENSDART00000170953 |
stoml2
|
stomatin (EPB72)-like 2 |
| chr4_+_70728926 | 0.38 |
|
|
|
| chr14_+_4071250 | 0.38 |
ENSDART00000162015
|
slc25a51b
|
solute carrier family 25, member 51b |
| chr18_+_40481566 | 0.38 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr15_+_26667763 | 0.37 |
ENSDART00000155352
|
ENSDARG00000074561
|
ENSDARG00000074561 |
| chr5_-_68765495 | 0.37 |
ENSDART00000097251
|
ENSDARG00000067564
|
ENSDARG00000067564 |
| chr5_-_68765567 | 0.37 |
ENSDART00000097251
|
ENSDARG00000067564
|
ENSDARG00000067564 |
| chr9_+_33247126 | 0.37 |
|
|
|
| chr22_-_6723168 | 0.37 |
|
|
|
| chr1_+_15928362 | 0.36 |
ENSDART00000149026
ENSDART00000159785 ENSDART00000164899 |
pcm1
|
pericentriolar material 1 |
| chr2_+_32760638 | 0.36 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr20_-_33887129 | 0.35 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr21_+_19283166 | 0.35 |
ENSDART00000063621
|
abraxas2a
|
abraxas 2a, BRISC complex subunit |
| chr25_+_28636948 | 0.35 |
|
|
|
| chr7_-_41446221 | 0.35 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr3_+_31530419 | 0.34 |
ENSDART00000113441
|
mylk5
|
myosin, light chain kinase 5 |
| chr3_+_22914688 | 0.34 |
ENSDART00000156472
|
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr5_-_54169271 | 0.34 |
ENSDART00000056213
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr16_-_34240818 | 0.34 |
ENSDART00000054026
|
rcc1
|
regulator of chromosome condensation 1 |
| chr1_+_51998508 | 0.33 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr23_-_32239909 | 0.33 |
|
|
|
| chr1_+_51998406 | 0.33 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr22_+_18218979 | 0.32 |
|
|
|
| chr2_+_52639159 | 0.32 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr8_-_54103762 | 0.32 |
|
|
|
| chr20_-_51843197 | 0.31 |
ENSDART00000129965
|
CABZ01079490.1
|
ENSDARG00000091595 |
| chr5_+_32751876 | 0.31 |
|
|
|
| chr6_-_33940201 | 0.30 |
ENSDART00000132762
|
akr1a1b
|
aldo-keto reductase family 1, member A1b (aldehyde reductase) |
| chr7_-_46940193 | 0.30 |
ENSDART00000161558
|
BX005073.2
|
ENSDARG00000099964 |
| chr15_+_28477893 | 0.30 |
ENSDART00000057257
|
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr21_-_43641419 | 0.30 |
ENSDART00000151486
ENSDART00000151115 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr15_+_6776788 | 0.30 |
ENSDART00000152653
|
ENSDARG00000096597
|
ENSDARG00000096597 |
| chr9_-_21427247 | 0.30 |
ENSDART00000146764
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr13_-_42598533 | 0.30 |
ENSDART00000160472
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr18_+_40481599 | 0.30 |
ENSDART00000098806
|
ugt5c3
|
UDP glucuronosyltransferase 5 family, polypeptide C3 |
| chr9_-_7308912 | 0.29 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr4_+_9477541 | 0.29 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr11_+_15743714 | 0.29 |
ENSDART00000167191
|
pank4
|
pantothenate kinase 4 |
| chr17_-_41000670 | 0.29 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr5_-_9443859 | 0.29 |
|
|
|
| chr18_-_2954048 | 0.29 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr5_-_979163 | 0.28 |
|
|
|
| chr10_+_4717721 | 0.28 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr23_+_17176587 | 0.28 |
ENSDART00000053414
|
commd7
|
COMM domain containing 7 |
| chr15_+_16950144 | 0.28 |
ENSDART00000049196
|
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr3_-_39163225 | 0.28 |
ENSDART00000102674
ENSDART00000167070 |
plcd3a
|
phospholipase C, delta 3a |
| chr1_-_55108003 | 0.27 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.5 | 1.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 1.5 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.4 | 1.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 1.4 | GO:0009223 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.3 | 3.5 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.2 | 0.7 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.2 | 1.0 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 0.9 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 1.0 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.2 | 0.6 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.2 | 0.5 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.4 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.2 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 2.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.3 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 4.4 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.7 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 1.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.2 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.8 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.2 | GO:0006895 | phagolysosome assembly(GO:0001845) Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.6 | GO:0060173 | limb development(GO:0060173) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.6 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.6 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 0.7 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 0.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.2 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.6 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 2.2 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 3.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.0 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.1 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.4 | 1.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.7 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.2 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.7 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 1.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 0.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 4.2 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 1.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 2.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.1 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.2 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.1 | 1.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 2.0 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.6 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |