Project
DANIO-CODE
Navigation
Downloads
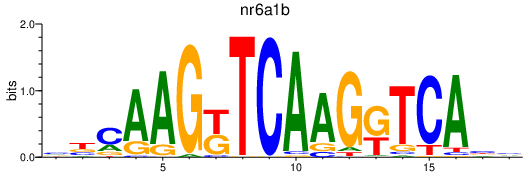
Results for nr6a1b
Z-value: 1.13
Transcription factors associated with nr6a1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
nr6a1b
|
ENSDARG00000014480 | nuclear receptor subfamily 6, group A, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| nr6a1b | dr10_dc_chr21_+_8106096_8106185 | -0.52 | 4.1e-02 | Click! |
Activity profile of nr6a1b motif
Sorted Z-values of nr6a1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of nr6a1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_30673985 | 3.85 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr25_+_7318688 | 3.85 |
ENSDART00000166496
|
cat
|
catalase |
| chr5_-_36503296 | 3.54 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr20_+_9141086 | 3.26 |
ENSDART00000064144
ENSDART00000144396 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr17_-_45021393 | 3.19 |
|
|
|
| chr2_-_50491234 | 3.06 |
ENSDART00000165678
|
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr25_+_17764396 | 2.78 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr2_-_22631167 | 2.67 |
ENSDART00000168653
ENSDART00000158558 |
fam110b
BX927330.1
|
family with sequence similarity 110, member B ENSDARG00000098356 |
| chr7_-_31346802 | 2.59 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr24_-_10253851 | 2.52 |
ENSDART00000127568
ENSDART00000106260 |
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr5_-_11471707 | 2.48 |
ENSDART00000166285
|
si:ch73-47f2.1
|
si:ch73-47f2.1 |
| chr8_-_11806698 | 2.32 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr4_+_9835529 | 2.31 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr7_-_31346844 | 2.28 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr18_+_8954323 | 2.25 |
ENSDART00000134827
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr8_+_25126238 | 2.17 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr8_-_11806733 | 2.07 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr17_+_18217416 | 1.92 |
ENSDART00000156616
|
BX465857.1
|
ENSDARG00000097921 |
| chr7_-_25252260 | 1.55 |
ENSDART00000173657
|
CU929416.1
|
ENSDARG00000105572 |
| chr11_+_44909949 | 1.47 |
ENSDART00000173116
ENSDART00000167011 ENSDART00000167226 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr14_+_29932533 | 1.46 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr15_-_16950049 | 1.42 |
|
|
|
| chr7_-_31346991 | 1.38 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr10_-_16910230 | 1.33 |
ENSDART00000171755
ENSDART00000170953 |
stoml2
|
stomatin (EPB72)-like 2 |
| chr9_-_7308912 | 1.28 |
ENSDART00000128352
|
mitd1
|
MIT, microtubule interacting and transport, domain containing 1 |
| chr2_-_56848527 | 1.21 |
ENSDART00000164330
|
CU634008.1
|
ENSDARG00000100430 |
| chr1_-_7882397 | 1.11 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr8_-_54103762 | 1.08 |
|
|
|
| chr23_-_32239909 | 1.07 |
|
|
|
| chr8_-_54103635 | 1.00 |
|
|
|
| chr4_+_9835639 | 1.00 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr1_+_16834347 | 0.97 |
ENSDART00000005593
ENSDART00000140076 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr18_-_2954175 | 0.94 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr12_-_18286637 | 0.93 |
ENSDART00000078767
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr5_-_36503236 | 0.90 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr2_+_38959335 | 0.89 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr3_-_9504876 | 0.88 |
ENSDART00000128731
|
rnps1
|
RNA binding protein S1, serine-rich domain |
| chr1_+_29058085 | 0.86 |
ENSDART00000133905
|
pcca
|
propionyl CoA carboxylase, alpha polypeptide |
| chr11_+_15743714 | 0.85 |
ENSDART00000167191
|
pank4
|
pantothenate kinase 4 |
| chr13_+_583640 | 0.84 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr7_-_7533176 | 0.84 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr21_-_32341308 | 0.83 |
ENSDART00000112550
|
mapk9
|
mitogen-activated protein kinase 9 |
| chr25_+_7318825 | 0.82 |
ENSDART00000159748
|
cat
|
catalase |
| KN149698v1_+_88937 | 0.82 |
ENSDART00000163341
|
ENSDARG00000100188
|
ENSDARG00000100188 |
| chr23_-_533016 | 0.82 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr10_-_16910257 | 0.82 |
ENSDART00000171755
ENSDART00000170953 |
stoml2
|
stomatin (EPB72)-like 2 |
| chr23_-_532974 | 0.82 |
ENSDART00000105316
|
samhd1
|
SAM domain and HD domain 1 |
| chr2_-_41888389 | 0.81 |
|
|
|
| chr15_+_26667763 | 0.77 |
ENSDART00000155352
|
ENSDARG00000074561
|
ENSDARG00000074561 |
| chr13_+_583744 | 0.75 |
ENSDART00000109737
|
cox15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr12_+_33294613 | 0.70 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr4_+_9477541 | 0.68 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr24_-_24999067 | 0.66 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr23_-_27647386 | 0.65 |
|
|
|
| chr19_+_24334831 | 0.55 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr24_+_24999138 | 0.55 |
ENSDART00000152095
ENSDART00000154988 ENSDART00000156098 |
CR383672.2
|
ENSDARG00000096452 |
| chr2_+_52639159 | 0.54 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr15_+_16950144 | 0.52 |
ENSDART00000049196
|
gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr17_-_41000670 | 0.52 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr12_-_18286666 | 0.49 |
ENSDART00000078767
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr16_+_27289443 | 0.49 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr16_+_27289472 | 0.48 |
ENSDART00000059013
|
sec61b
|
Sec61 translocon beta subunit |
| chr18_-_2954048 | 0.43 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr5_-_61644464 | 0.43 |
ENSDART00000168993
|
spata22
|
spermatogenesis associated 22 |
| chr19_-_6503643 | 0.42 |
|
|
|
| chr11_+_44805115 | 0.36 |
ENSDART00000167828
|
tmc6b
|
transmembrane channel-like 6b |
| chr16_+_45726666 | 0.35 |
ENSDART00000060809
|
smg5
|
SMG5 nonsense mediated mRNA decay factor |
| chr1_-_45941649 | 0.33 |
ENSDART00000148893
|
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr9_+_31469848 | 0.33 |
|
|
|
| chr3_-_40834641 | 0.29 |
ENSDART00000018676
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr16_+_14326410 | 0.28 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr19_+_24334731 | 0.17 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr19_+_5218562 | 0.14 |
ENSDART00000151310
|
si:dkey-89b17.4
|
si:dkey-89b17.4 |
| chr22_+_18218979 | 0.14 |
|
|
|
| chr24_-_22369651 | 0.14 |
|
|
|
| chr3_-_13449591 | 0.09 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr10_+_41237437 | 0.09 |
ENSDART00000141657
|
anxa4
|
annexin A4 |
| chr6_-_30283366 | 0.09 |
|
|
|
| chr2_-_59019569 | 0.03 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.8 | 3.3 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.5 | 1.6 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) purine deoxyribonucleotide catabolic process(GO:0009155) purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.4 | 4.7 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.3 | 1.0 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.3 | 3.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) mitotic DNA replication(GO:1902969) |
| 0.2 | 1.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.2 | 3.3 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.1 | 2.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.5 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.0 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.9 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 4.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 2.8 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.0 | 0.8 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.4 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 2.2 | GO:0007188 | G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger(GO:0007187) adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 3.3 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 0.9 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 3.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 4.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 3.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.8 | 3.1 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.5 | 1.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 1.5 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.3 | 2.7 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.2 | 0.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 4.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 1.6 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.1 | 3.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 4.4 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 0.9 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.3 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.7 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 1.0 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.1 | 0.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.2 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.6 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.4 | GO:0030276 | clathrin binding(GO:0030276) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 3.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 2.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 0.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 4.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.8 | 3.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.7 | 3.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.4 | 2.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.3 | 3.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 0.8 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.9 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 1.4 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |